Figure 1. Structure of Rab‐specific PPM1H phosphatase.
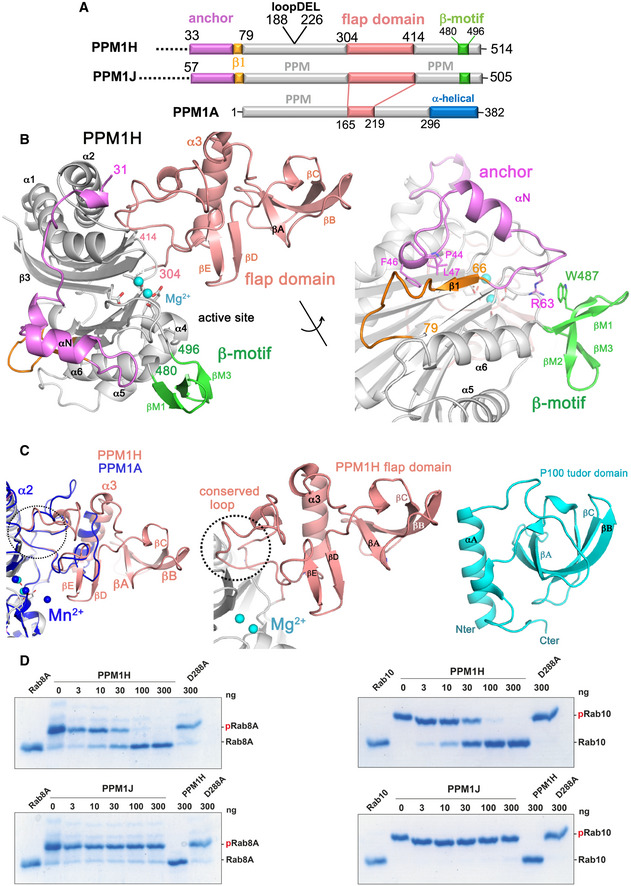
- Domain organization of PPM1H, PPM1J and PPM1A. The annotation of regions (anchor, flap domain) is discussed in the text. The loop deletion (188–226) that was engineered to improve diffraction is indicated.
- Ribbon model of the enzyme with a view to the catalytic cleft that contains two Mg2+ ions (cyan spheres). The N‐terminal region is magenta (33–71), the flap domain is a wheat colour, and the β‐sheet motif is green. The loop deletion (188–226) connects α1/α2 on the opposite face relative to the active site. The back view of the enzyme is also shown with a 180° rotation around the axis indicated. Parts of the anchor (RPxFL motif, magenta) that interact with the globular core are shown as stick models, and discussed in the text. The β1 strand is orange to emphasize its non‐canonical conformation due to the presence of the preceding anchor.
- Comparisons of the flap domain of PPM1H with PPM1A (left) and the tudor domain (right). Apart from a conserved loop (dotted circle) which forms an interface with the catalytic domain, the sequences and structures of flaps are diverse among the PPM family.
- The indicated amounts of recombinant wild‐type and mutant PPM1H or PPM1J (with a His‐Sumo N‐terminal tag, expressed in E. coli) were incubated in vitro with 2.5 µg pThr72‐phosphorylated Rab8a (left) or pThr73‐phosphorylated Rab10 (right) for 20 min in the presence of 10 mM MgCl2 in 40 mM HEPES pH 7.5 buffer. Reactions were terminated by addition of SDS sample buffer and analysed by Phos‐tag gel electrophoresis that separates phosphorylated (slow migrating) and dephosphorylated Rabs. The gel was stained with Instant Blue Coomassie. D288A substrate‐trapping (inactive) variant of PPM1H was used as a control.
