Figure EV1. The “loop deletion” variant of PPM1H is active in vitro and in cells.
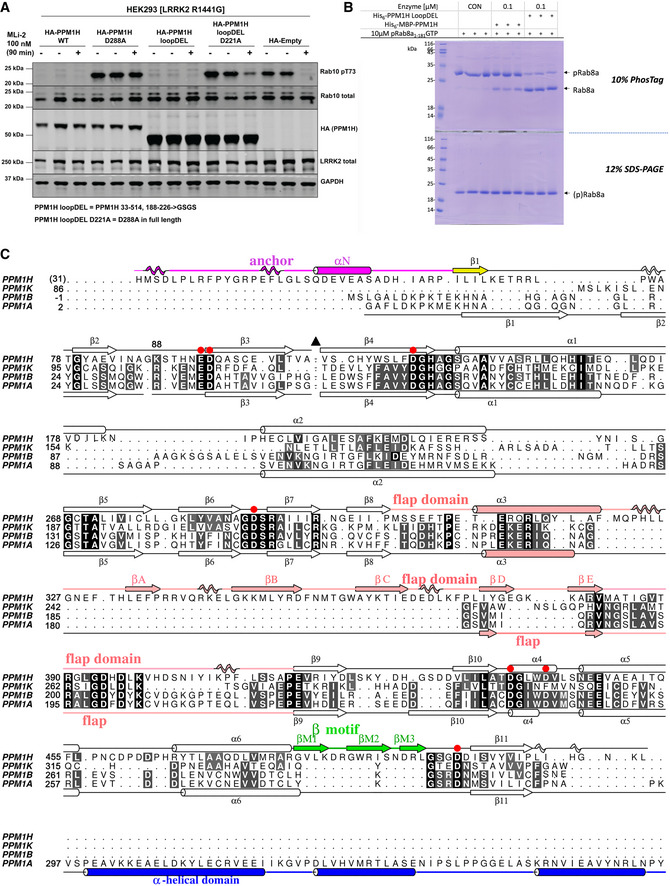
- HEK293 cells overexpressing indicated constructs were treated ± 200 nM MLi‐2 for 90 min and then lysed. 10 μg whole cell lysate was subjected to immunoblot analysis with the indicated antibodies at 1 μg/ml final concentration, and membranes were analysed using the OdysseyClx Western blot imaging system. Each lane represents cell extract obtained from a different dish of cells (two replicates per condition without MLi‐2 treatment, one replicate per condition with MLi‐2 treatment). The “loopDEL” variant is the segment 33–514 with the region 188‐226 replaced by the sequence “GSGS”.
- In vitro assay of PPM1H activity using a PhosTag gel. Substrate pRab8a (GTP form, 10 μg) was incubated with WT PPM1H ± loopDEL for 15 min at room temperature. The full‐length variant was fused to maltose‐binding protein (MBP). The “loopDEL” variant was from 33 to 514 and was used for crystallization studies. A conventional 12% SDS–PAGE is shown in parallel lanes below the PhosTag gel.
- Structure‐based sequence alignment of human PPMs from their associated PDB files using Chimera software (Pettersen et al, 2004). The PDB codes are 4ra2 (PPM1A; (Pan et al, 2015)), 2p8e (PPM1B; (Almo et al, 2007)) and 2iq1 (PPM1K; (Almo et al, 2007)). Residue His31 of PPM1H is in brackets since the first two residues (His‐Met) arise from a cloning artefact. Secondary structures of PPM1H and PPM1A are above and below the sequences, respectively. Colours of secondary structures correspond to the scheme in Fig 1. The black triangle is a loop in PPM1H (188‐226) that has been removed to simplify the alignment. The C‐terminal α‐helical domain of PPM1A is blue. Red circles are aspartate residues that directly coordinate metal ions.
