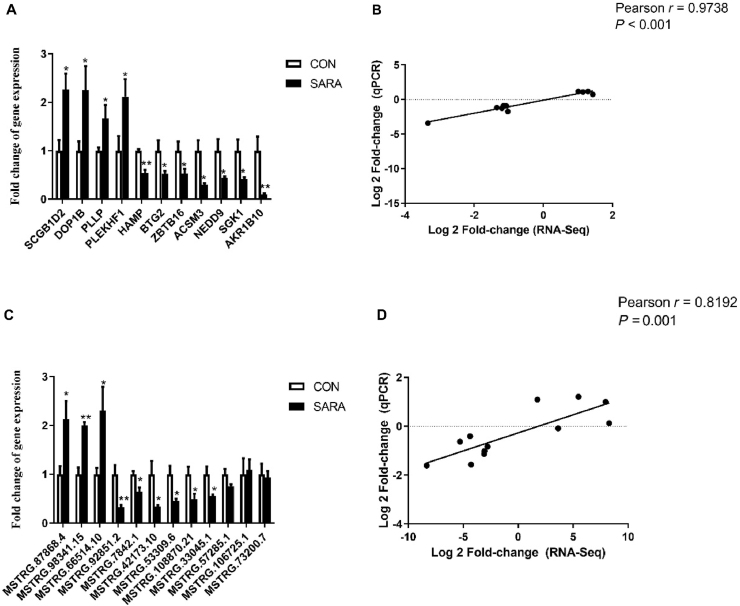
Fig. 7.
qPCR validation of RNA-seq results. (A) Expression of mRNA in the liver. (B) Pearson correlation analysis for mRNA between RNA-seq and qPCR. Fold change indicates the ratio of average expression of SARA-affected mRNA relative to controls. (C) Expression of lncRNA in the liver. (D) Pearson correlation analysis for lncRNA between RNA-seq and qPCR. Fold change indicates the ratio of average expression of SARA-affected lncRNA relative to controls. Fold difference for each gene was calculated via the FPKM values of CON and SARA. GAPDH was used as the reference gene for genes expression. The data were analyzed by an independent-samples t-test using the Compare Means of SPASS 11.0 for Windows (StaSoft Inc, Tulsa, OK, USA). Values are mean ± SEM. ∗P < 0.05, ∗∗P < 0.01 (CON vs SARA). qPCR = quantitative PCR; FPKM = fragments per kilo-base of exon per million fragments; CON = control; SARA = subacute ruminal acidosis; GAPDH = glyceraldehyde-3-phosphate dehydrogenase.