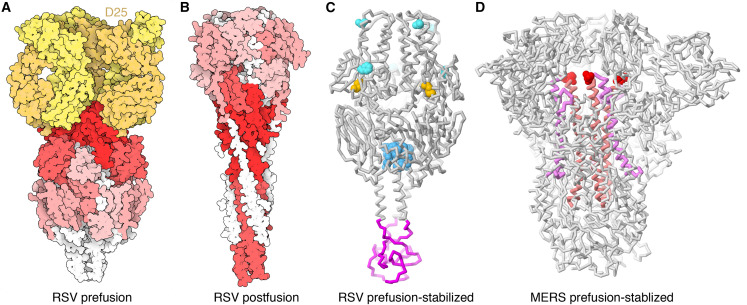
Figure 4.
Stabilization of prefusion conformations of viral surface glycoproteins
(A) Space-filling representation of the prefusion conformation of the RSV glycoprotein F ectodomain (PDB: 4jhw; McLellan et al., 2013b), with antibody epitopes colored by neutralization sensitivity (dark red, highest; light red, intermediate; pink, lowest; white, antibody inaccessible) and a neutralizing antibody D25 Fab shown in yellow.
(B) Space-filling representation of the post-fusion conformation of the RSV glycoprotein F ectodomain (PDB: 3rrr; McLellan et al., 2011), showing that the most neutralization-sensitive epitopes are inaccessible to antibodies in the altered conformation.
(C) Polypeptide chain backbone representation of the RSV F glycoprotein stabilized in the prefusion conformation (PDB: 4mmv; McLellan et al., 2013a) with an engineered disulfide bridge (yellow), several sites of mutation to fill pockets (turquoise and blue), and a foldon to stabilize the homotrimer (magenta).
(D) Polypeptide chain backbone representation of the MERS-CoV virus spike protein homotrimer stabilized in a prefusion conformation (PDB: 5w9j; Pallesen et al., 2017) with two prolines (red spheres) linking the heptad repeat (HR1, magenta) and the central helix (pink). (A) and (B) created with Illustrate software; (C) and (D) created with Mol∗ (Sehnal et al., 2021) at the RCSB PDB website.