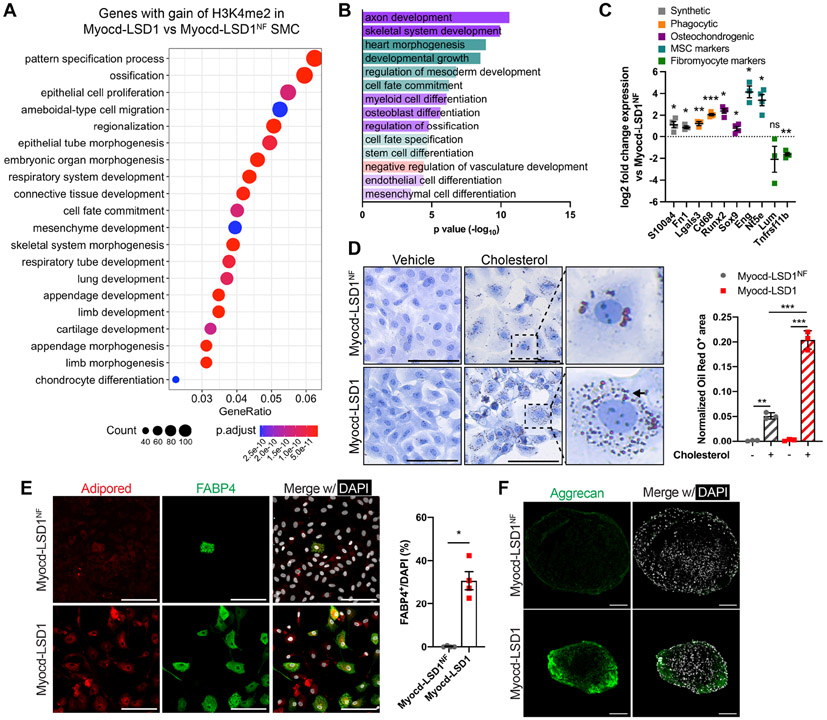
Figure 6: Loss of H3K4me2 on myocardin-regulated genes exacerbates SMC plasticity.
A. Top20 Gene Ontology pathways of genes with enhanced H3K4me2 enrichment in Myocd-LSD1 SMC compared with Myocd-LSD1NF SMC. B. Significantly upregulated GO pathways related with lineage plasticity from RNAseq in Myocd-LSD1 vs Myocd-LSD1NF SMC. C. Expression of markers of SMC phenotypic transitions in Myocd-LSD1 SMC. D. Oil Red O staining in Myocd-LSD1 and Myocd-LSD1NF SMC treated with cholesterol (40 μg/ml) or vehicle for 48h and quantification of Oil Red O+ area normalized to cellular area. Arrow: example of intracellular Oil Red O+ lipid vacuoles. Scale bar: 100μm. E. FABP4 and lipid droplets staining (left) and quantification of FABP4+ SMC (right) in Myocd-LSD1 and Myocd-LSD1NF SMC cultured in adipogenic differentiation media. Scale bar: 100μm. F. Immunofluorescent staining with Aggrecan in SMC cultured in chondrogenic differentiation media. Scale bar: 100μm. Data are represented as mean ± s.e.m of 3-4 independent experiments. Groups were compared by One-Way ANOVA (D) and Student t-test (C, E). * p<0.05, ** p<0.001, *** p<0.0001.