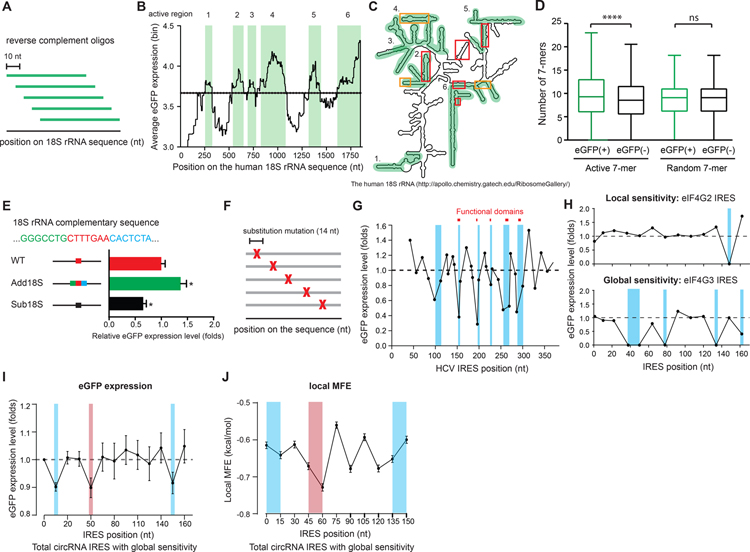
Figure 3. 18S rRNA complementary sequence on the IRES facilitates circRNA translation.
(A) A schematic of the sliding-window approach for mapping the active regions on the human 18S rRNA. (B) Quantification of the mean eGFP expression of the oligos overlapping with the corresponding position across the human 18S rRNA. Dashed line: background eGFP expression. Green shaded: active regions on the 18S rRNA. (C) An illustration of the secondary structure of human 18S rRNA showing the active regions (green) and reported mRNA (red) or IRES RNA (orange) contact regions. (D) Quantification of the number of the 18S rRNA active 7-mers or the random 7-mers harbored by eGFP(+) or eGFP(−) oligos plotted on a Tukey box-plot. (E) Quantification of the IRES activity for the oligo with higher or lower 18S rRNA complementarity. Error bar: SEM. (F) A schematic of the synthetic oligos for systematic scanning mutagenesis. (G) The eGFP expression of each oligo containing the random substitution mutation at the corresponding position on HCV IRES. The eGFP expression for each oligo was normalized to the mean eGFP expression of all the oligos on the HCV IRES. (H) Examples of circRNA IRES with local and global sensitivity identified by scanning mutagenesis. Blue shaded: the identified essential elements on the IRES. (I) The mean eGFP expression of all the circRNA IRES oligos with global sensitivity at each mutation position across the IRES (blue: 5–15 nt and 135–165 nt; red: 40–60 nt). (J) Quantification of the local MFE in a 15 nt sliding window on the IRES (blue: 5–15 nt and 135–165 nt; red: 40–60 nt).