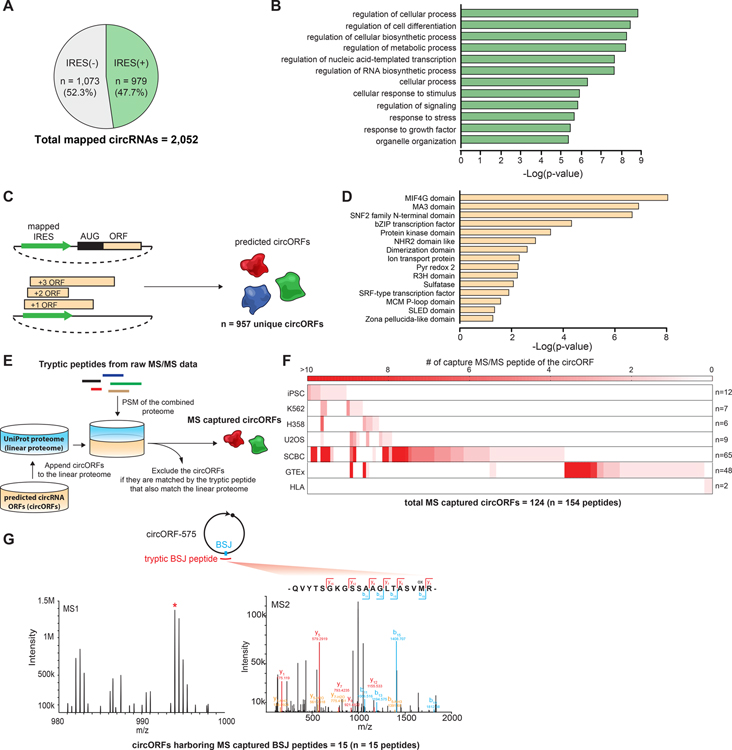
Figure 6. Identification of putative endogenous circRNA-encoded proteins.
(A) Quantification of the percentage of IRES-mapped human endogenous circRNAs. (B) Top 12 represented biological processes from GO term analysis that are enriched in the parent genes of IRES(+) circRNAs. (C) A schematic of generating the putative endogenous circORF list. (D) Top 15 represented conserved motifs from Pfam analysis that are enriched in the predicted circORFs. (E) A schematic of peptidomic validation of the putative circORFs. (F) The heat map showing the number of the unique MS/MS peptides detected in different peptidomic datasets for each of the peptidomic detected circORF. The number on the right indicates the total number of different circORFs detected in the corresponding dataset. (G) The MS1 and MS2 spectra of a representative tryptic BSJ peptide captured from circORF_575.