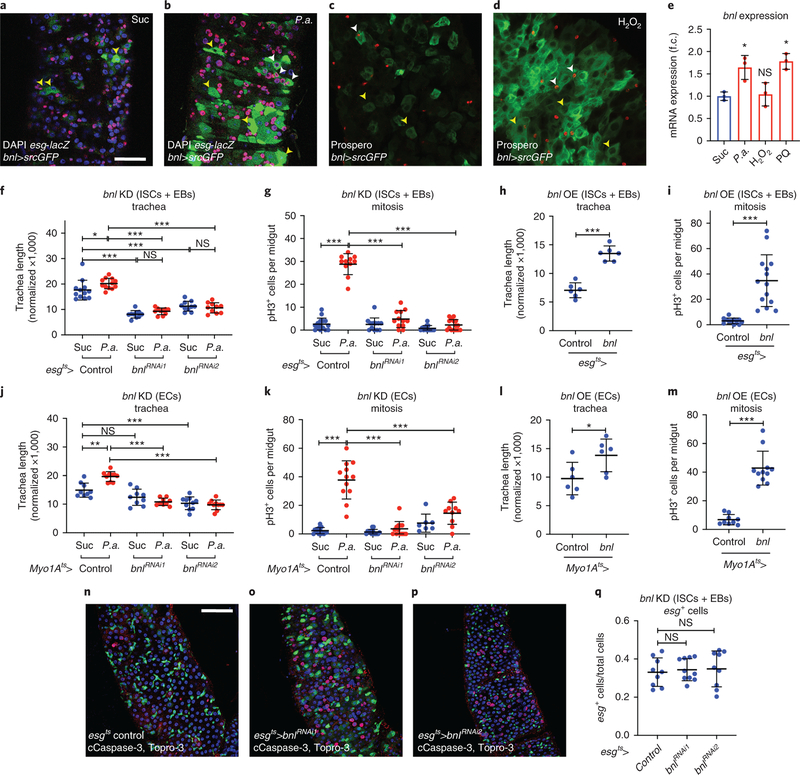
Figure 2. The FGF/Bnl is expressed in the midgut epithelium and controls ISC proliferation and TTC remodelling.
a-b. Control (a) and P.a.-infected (b) bnl-Gal4>UAS-srcGFP midguts (R5 region) with esg-lacZ-labeled (red) ISCs and EBs, and DAPI (blue) staining all the nuclei. c-d. Control (c) and H2O2-fed (d) bnl-Gal4>UAS-srcGFP midguts (R2 region) with Prospero-labeled (red) EEs, and DAPI (blue) staining all the nuclei. Yellow arrowheads indicate bnl-positive ECs (big cells) and white arrowheads indicate bnl-positive esg-positive progenitors (a-b) and EEs (c-d). e. RT-qPCR for bnl mRNA expression in w1118 whole adult midguts upon different stresses. The average and standard deviation of n=3 biological expreriments are plotted. f-g. Quantification of TTC branching (f, n=12,12,10,10,10,10) and mitosis (g, n=15,11,12,12,13,15) upon progenitor-specific bnl knockdown with or without P.a infection. h-i. Quantification of TTC branching (h, n=6 each) and mitosis (i, n=12,14) upon progenitor-specific bnl overexpression. j-k. Quantification of TTC branching (j, n=9,10,9,9,10,10) and mitosis (k, n=15,12,13,13,7,10) upon EC-specific bnl knockdown with or without P.a infection. l-m. Quantification of TTC branching (l, n=6 each) and mitosis (m, n=9,11) upon EC-specific bnl overexpression. n-p. Midguts (R5 region) of control (n) and esgts-Gal4 progenitor-specific bnl knockdown (o-p), stained for ISCs+EBs (green), cleaved Caspase-3 (red) and Topro-3 (blue). q. Quantification of esg-positive ISCs+EBs in control and progenitor-specific bnl knockdown (n=9,10,9). Scale bars: 37.5 μm in a-d, 75 μm in n-p. Data are presented as mean values ± SD. Statistical significance (t-tested, two-sided): ns, not significant, * 0.01<p≤0.05, ** 0.001<p≤0.01 and *** p≤0.001.