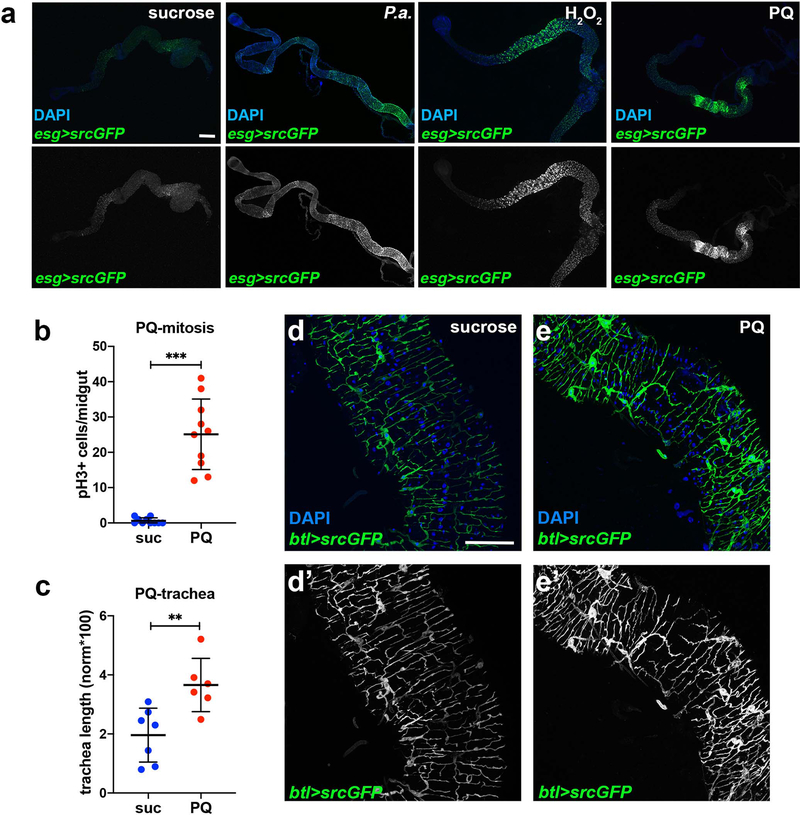
Extended Data Fig. 1. Infection and oxidative damage increase esg>GFP+ cells in the midgut and associate with increased TTC branching.
a. Adult midgut intestinal progenitors labeled with esgNP5130-Gal4>UAS-srcGFP were imaged in unchallenged conditions (4% sucrose) and upon oral P.a. infection (48hrs), and feeding with H2O2 (48hrs) and PQ (24hrs). DAPI (blue) in the upper panels stains all midgut nuclei. The bottom panels show the GFP-labeled progenitors separately. P.a. and PQ expanded the intestinal progenitors with a posterior midgut bias, whereas H2O2 exhibited an anterior midgut bias. b-c. Quantification of midgut mitosis (b, n=10 each) and TTC branching (c, n=7,6) in PQ-treated flies. d-e. Posterior midgut (R4) images of btl-Gal4>UAS-srcGFP flies in baseline conditions (sucrose) and upon PQ feeding. DAPI (blue) staining all the nuclei. Single channel images of the GFP are shown in d’-e’. f-g. Posterior midgut images of QF6>QUAS-mtdTomato flies in baseline conditions exhibit tracheal expression of the reporter. Midgut epithelial ECs with low expression of the reporter are visible is zoomed image (g). Single channel images of the Tomato are shown in f’-g’. Scale bars: 300 μm in a, 75 μm in d-g. Data are presented as mean values ± SD. Statistical significance (t-tested, two-sided for b, and U-tested for c): ns, not significant, * 0.01<p≤0.05, ** 0.001<p≤0.01 and *** p≤0.001.