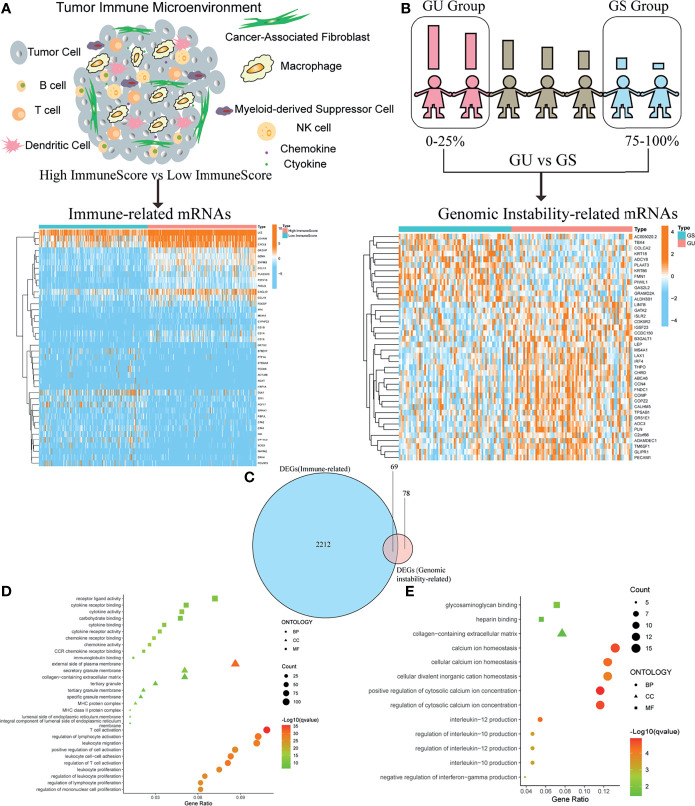
Figure 2.
Identification of differentially expressed genes (DEGs) both correlated with tumor immune microenvironment (TIME) and genomic instability of ovarian cancer (OC). (A) Heatmap for immune-related DEGs generated by comparison of the high score group vs. the low score group in ImmuneScore. The row name of heatmap is the gene name, and the column name is the ID of samples that are not shown in the plot. Differentially expressed genes were determined by Wilcoxon rank sum test with q < 0.05 and fold-change > 0.58 after log2transformation as the significance threshold. (B) Heatmap for genomic instability-related DEGs by comparison of the genomic unstable (GU) group vs. the genomic stable (GS) group, similar to (A). (C) Venn plot showing common DEGs shared by immune-related DEGs and genomic instability-related DEGs, and q < 0.05 and fold-change > 0.58 after log2transformation as the DEGs significance filtering threshold. (D) GO enrichment analysis for 2,281 immune-related DEGs; terms with p and q < 0.05 were believed to be enriched significantly. (E) GO enrichment analysis for 147 genomic instability-related DEGs; terms with p and q < 0.05 were believed to be enriched significantly.