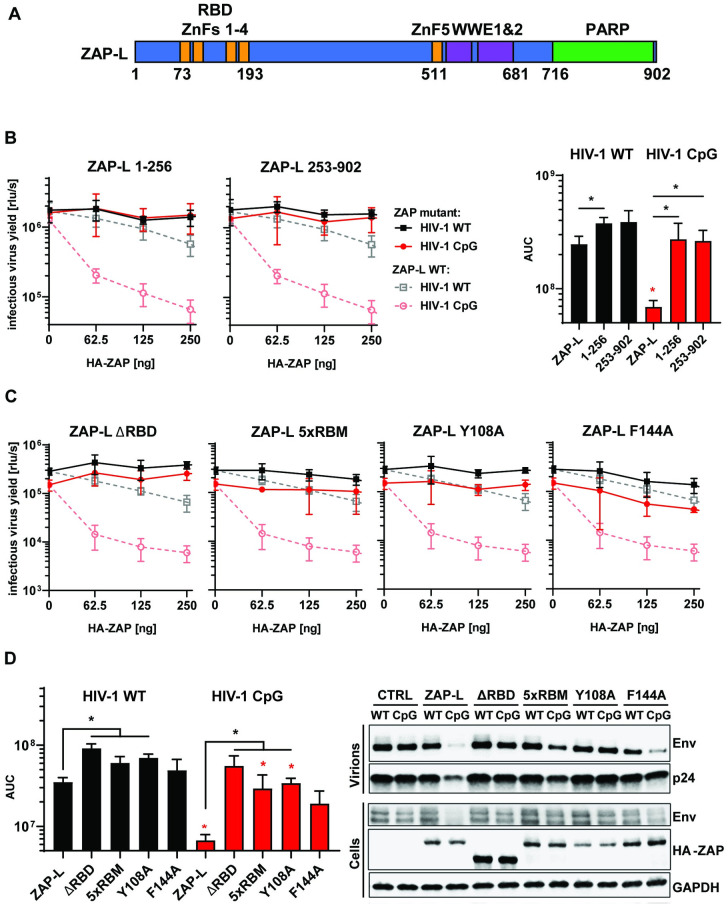
Fig 1. RNA binding is crucial for ZAP’s antiviral activity.
(A) Schematic showing domain organisation of long isoform of ZAP (ZAP-L): four N-terminal zinc fingers form RNA binding domain (RBD), fifth zinc finger (ZnF5) and two WWE domains are located in the central part and catalytically inactive Poly(ADP-ribose) polymerase (PARP) domain is at the C-terminal part. (B) Infectious virus yield measured by TZM-bl infectivity assay in relative light units per second [rlu/s] obtained from HEK293T ZAP KO cells co-transfected with wild type (WT; black) HIV-1 and CpG-enriched mutant (CpG-high; red) viruses and increasing doses of pcDNA HA-ZAP constructs encoding the full-length ZAP-L (dashed line), 1-256aa or 253-902aa parts of the protein (solid lines) (left panel). Area Under the Curve (AUC) calculated from the same titration experiments (right panel). (C) Infectious virus yield from HEK293T ZAP KO co-transfected with WT and mutant virus and increasing concentration of pcDNA HA-ZAP with truncated ZAP 253–902 (ΔRBD), ZAP-L mutant unable to bind RNA (V72A/Y108A/F144A/H176A/R189A; 5xRBM) or ZAP-L with substitutions at positions in direct contact with bound RNA CpG (Y108A and F144A) and (D, left panel) derived AUC values. (right panel) representative western blot of produced virions and ZAP transfected (250ng) cells showing viral Env and Gag (p24) proteins as well as HA-tagged ZAP and GAPDH loading control. Mean of n = 3 +/- SD. * p<0.05 for HIV-1 CpG compared to HIV-1 WT for the same ZAP construct. * p < 0.05 for the comparisons demarked by the lines.