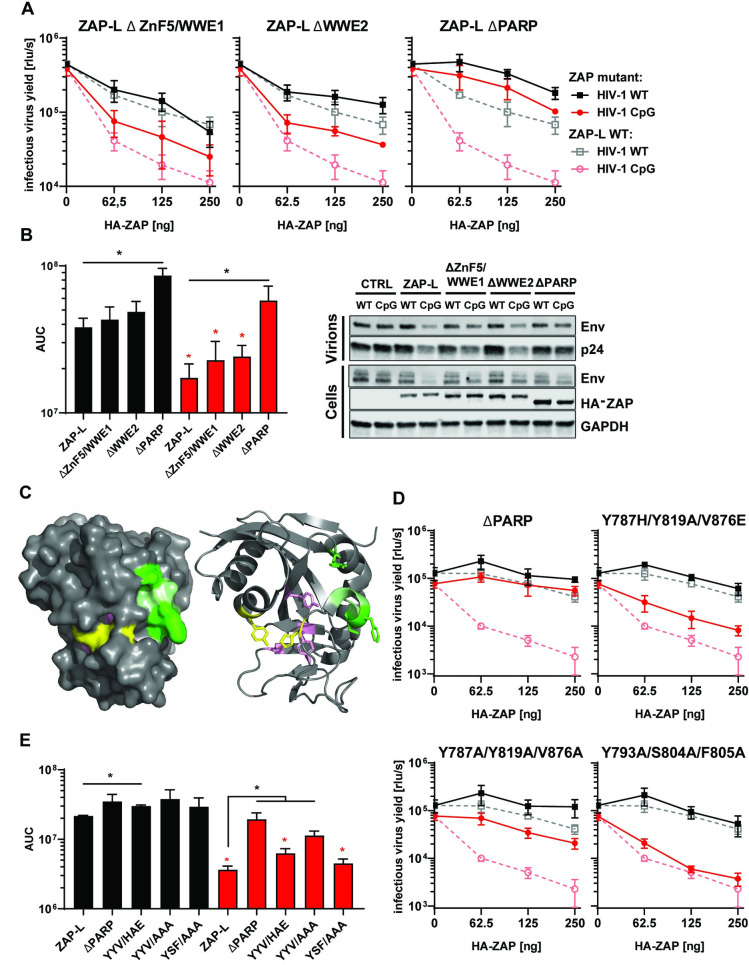
Fig 2. Determinants of ZAP’s function located outside RBD.
(A) Infectious virus yield from HEK293T ZAP KO co-transfected with WT (black) and mutant (red) virus and increasing concentration of pcDNA HA-ZAP-L control (dashed line) or mutated pcDNA HA-ZAP with deleted ZnF5 and first WWE domain (Δ511–563; ΔZnF5/WWE1), second WWE (Δ594–681; ΔWWE2) or PARP domain (Δ716–902; ΔPARP). (B) Corresponding AUC values and representative western blot (250ng). (C) Position of studied residues in crystal structure of ZAP’s PARP domain. Residues under positive selection in primates are shown in green, canonical triad positions in pink and residues forming the salt bridge which closes the NAD+ binding grove are shown in yellow. (D) Infectious virus yield from HEK293T ZAP KO co-transfected with WT (black) and mutant (red) virus and increasing concentration of pcDNA HA-ZAP-L control (dashed line), missing PARP domain or carrying amino acid substitutions in alternate triad motif (Y786H/Y818A/V875E, Y786A/Y818/V875A), or residues under positive selection (Y793A/S804A/F805A) (solid lines). (E) Corresponding AUC values. Mean of n = 3 +/- SD. * p<0.05 for HIV-1 CpG compared to HIV-1 WT for the same ZAP construct. * p < 0.05 for the comparisons demarked by the lines.