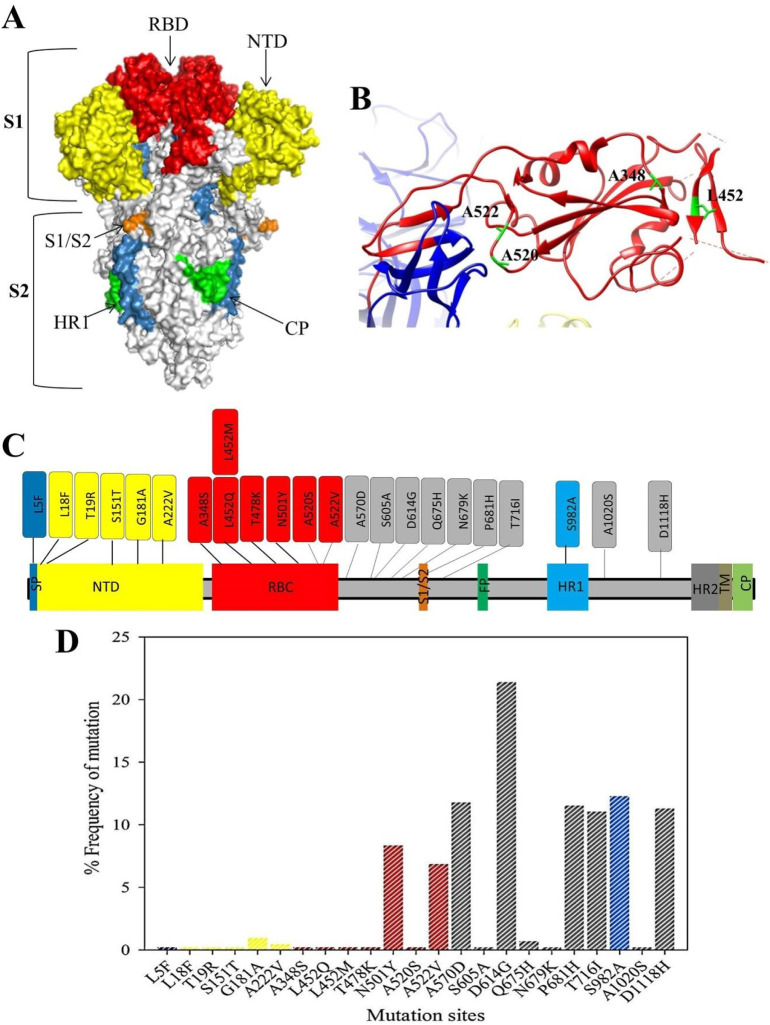
Fig. 4.
Structure and the frequency of the mutations in the spike protein region among SARS-COV2 strains from Iraq. (A) Trimer structure of SARS-COV2 protein (PDB: 6VXX), the RBD (residue 319–541) is coloured red. (B): The ribbon structure of RBD of the monomeric structure of the protein and the locations of 5 mutations in the region of the protein. (C) Schematic representation of the different regions of SARS-COV2 and approximate mutations in each of the regions. SP: signal peptide; NTD: N-terminal domain; RBD: Ribosomal binding domain; S1/ S2 protease cleavage site; FP: fusion peptide; HR1: heptad repeat 1; HR2: heptad repeat 2; TM: transmembrane domain; CP: cytoplasmic peptide. (D) Percentage of the mutations of each of the residues located in the Spike proteins among 91 Iraqi isolates. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)