Figure 1. The VP40 ring binds the 3′ untranslated region (UTR) of cellular mRNA.
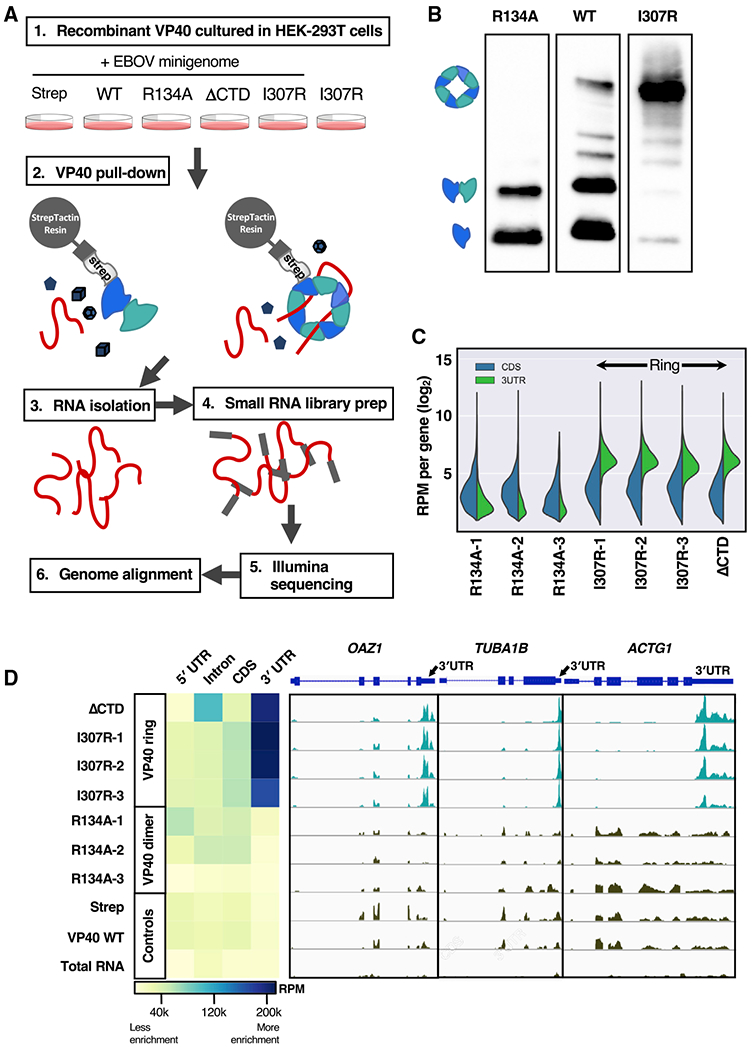
(A) HEK293T cells were transfected with Strep-tagged VP40-WT, three VP40-R134A biological replicates, VP40ΔCTD, three VP40-I307R biological replicates, and Strep-tag alone as a control. An Ebola minigenome replicon system was co-transfected in all cases, with the exception of one I307R biological replicate (I307R-3) as a control. The VP40 was pulled down from the cellular lysate by Strep-Tactin resin, after which the RNA was extracted from each VP40 sample and deep sequenced.
(B) Three of our Strep-Tactin purified VP40 samples run in a blue native protein gel and blotted for Strep-tag. The display as separate images is solely to remove irrelevant lanes. We identify the dominant protein bands seen here as the VP40 monomer, dimer, and octameric ring.
(C) For seven of our RNA samples, the number of reads that align with the coding sequence (CDS) or with the 3′ UTR, respectively, of each of the 1,986 cellular mRNAs that are highest expressed in our data. We observe 3′ UTR enrichment in the ring-forming VP40 mutant samples (VP40-I307R biological replicates, VP40ΔCTD). Read counts were normalized to reads per million (RPM).
(D) For each of our samples, the heatmap displays the RNA RPM that align with each mRNA component (5′ UTR, intron, CDS, or 3′ UTR). For the genes OAZ1, TUBA1B, and ACTG1, we also display the per-gene read peaks. We observed 3′ UTR enrichment in the ring-forming VP40 mutant samples (VP40-I307R biological replicates, VP40ΔCTD). An Ebola minigenome replicon system was co-transfected in all cases except I307R-3 and total RNA (RNA extracted from the HEK293T cellular lysate).
