Figure 2. The VP40 ring’s RNA binding preference is specific to the mRNA 3′ UTR.
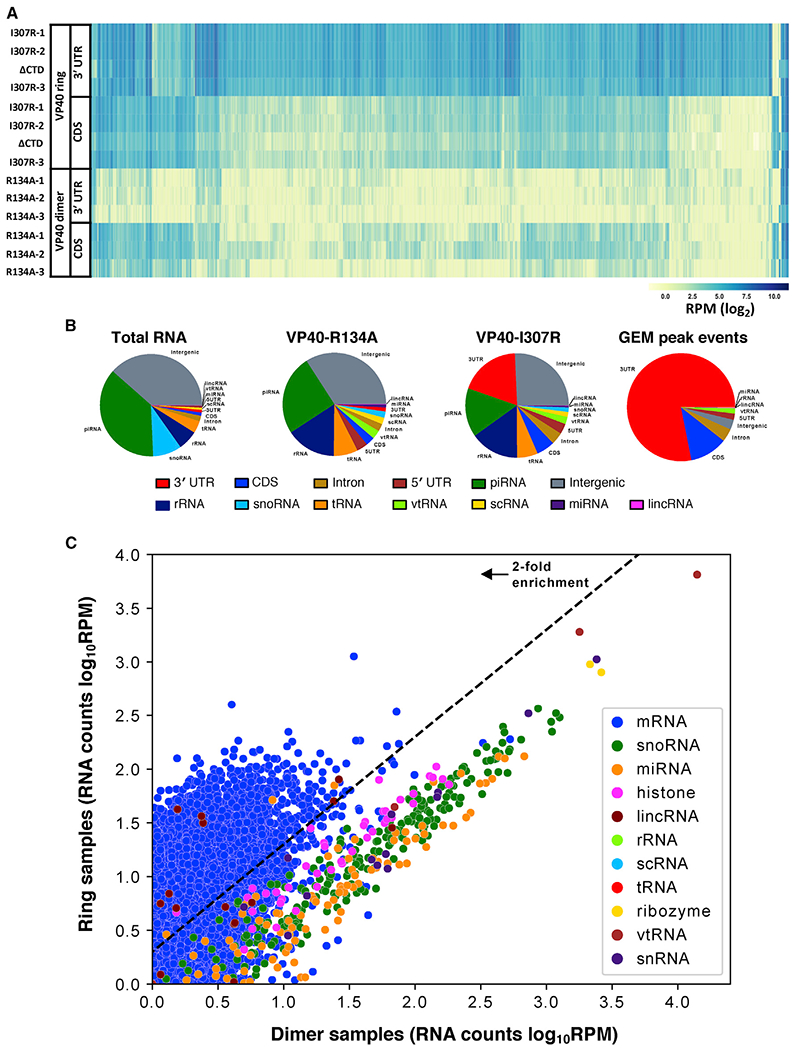
(A) The number of reads that align with the 3′ UTR and with the CDS, respectively, of each of the 1,986 cellular mRNAs that are highest expressed in our data. We observed that the ring-formingVP40 mutant samples (VP40-I307R biological replicates, VP40ΔCTD) have more 3′ UTR reads than CDS reads, while for the dimer-forming VP40 mutant samples (VP40-R134A biological replicates) the opposite is true. Read counts were normalized to RPM.
(B) Reads categorized by RNA feature (the components of mRNA including the 3′ UTR, the CDS, the introns, and the 5′ UTR; also, several forms of non-coding RNA) for a ring-forming VP40 mutant sample (biological replicate VP40-I307R-1) and two non-ring-forming samples (biological replicate VP40-R134A-1, total RNA control). Also included is a similar categorization of the VP40-RNA binding event loci called by the GEM peak-finding algorithm (Figure 3). See also Figure S1.
(C) The cellular RNA transcript abundances in our VP40 samples, comparing our ring-forming VP40 mutant samples versus our dimer-forming VP40 mutant samples. Reads were aligned with the human genome (hg19), filtered to remove duplicates and low-quality alignments, and normalized to RPM. We then averaged over our three VP40-I307R biological replicates and one VP40ΔCTD sample to determine the ring samples value, and over our three VP40-R134A biological replicates to determine the dimer samples value. 7,072 genes are plotted, representing all UCSC Known Genes for which both averages are at least 1 RPM. See also Figure S2.
