Figure 3. Analysis reveals a guanine- and adenine-rich 3′ UTR binding motif.
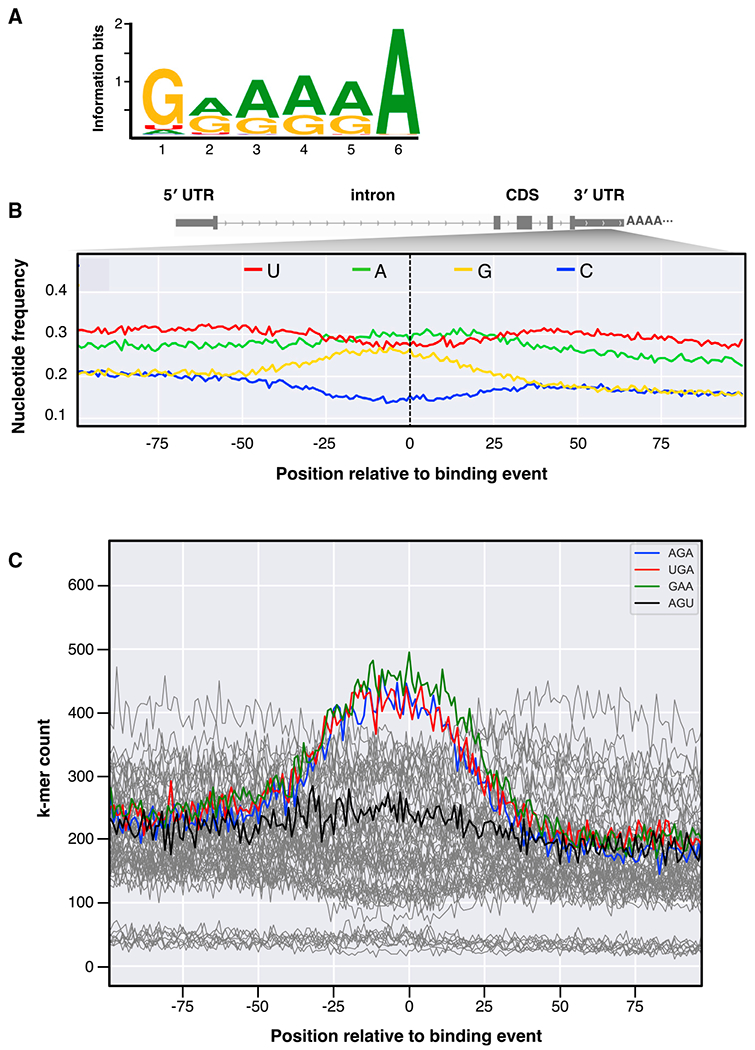
(A) The highest ranked binding motif called by the genome-wide event finding and motif discovery (GEM) analysis of the VP40 binding events in mRNA 3′ UTRs. Letter heights indicate the nucleotide base frequencies (guanine, yellow; adenine, green; uracil, red; cytosine, blue).
(B) The average nucleotide base frequencies within 100 nt of the genome positioning system (GPS)-identified binding events in 3′ UTRs. Human 3′ UTRs are on average U/A-rich, G/C-poor (left), but G/A enrichment extends ~25–50 nt to either side of the binding events (center).
(C) For each possible 3-nt k-mer (with the data for the k-mers AGA, UGA, GAA, and AGU highlighted), the number of times the k-mer occurs in the human genome at the specified offset relative to any GPS-identified 3′ UTR binding event.
