Figure 7. The hypothesized interaction of an mRNA 3′ UTR with VP40 dimers to generate the VP40 ring.
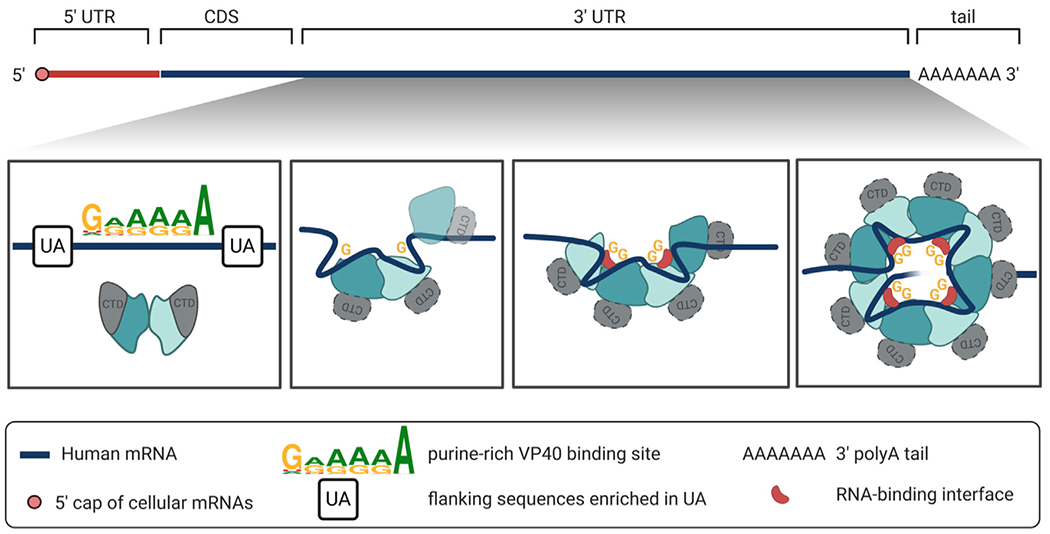
Top: a human mRNA comprises a 5′ UTR, a CDS, a 3 UTR, and a polyadenylate tail. First panel: as long as the VP40 remains in its butterfly-shaped dimer configuration, it does not bind RNA. Second panel: however, when a VP40 dimer encounters suitable RNA, namely a G/A-rich binding sequence within the generally U/A-rich mRNA 3′ UTR, the dimer interface is disrupted, as well as the interface between the NTD and the CTD of each VP40 protomer. The protein-protein interfaces are rearranged, while the RNA remains bound to each NTD via a central guanine base. Third panel: the 3′ UTR strand may now assist in bringing rearranged VP40 dimers together. A half-ring may form as a transformation intermediate. Fourth panel: the octameric VP40 ring is assembled. A single 3′ UTR strand may loop through the ring to bind at the eight known binding sites, two at each of the ring’s four RNA binding clefts.
