Abstract
Purpose:
Artificial intelligence diagnosis and triage of large vessel occlusion may quicken clinical response for a subset of time-sensitive acute ischemic stroke patients, improving outcomes. Differences in architectural elements within data-driven convolutional neural network (CNN) models impact performance. Foreknowledge of effective model architectural elements for domain-specific problems can narrow the search for candidate models and inform strategic model design and adaptation to optimize performance on available data. Here, we study CNN architectures with a range of learnable parameters and which span inclusion of architectural elements, such as parallel processing branches and residual connections with varying methods of recombining residual information.
Methods:
We compare five CNNs: ResNet-50, DenseNet-121, EfficientNet-B0, PhiNet, and an Inception module-based network, on a computed tomography angiography large vessel occlusion detection task. The models were trained and preliminarily evaluated with 10-fold cross-validation on preprocessed scans (n=240). An ablation study was performed on PhiNet due to superior cross-validated test performance across accuracy, precision, recall, specificity, and F1 score. The final evaluation of all models was performed on a withheld external validation set (n=60) and these predictions were subsequently calibrated with sigmoid curves.
Results:
Uncalibrated results on the withheld external validation set show that DenseNet-121 had the best average performance on accuracy, precision, recall, specificity, and F1 score. After calibration DenseNet-121 maintained superior performance on all metrics except recall.
Conclusions:
The number of learnable parameters in our five models and best-ablated PhiNet directly related to cross-validated test performance—the smaller the model the better. However, this pattern did not hold when looking at generalization on the withheld external validation set. DenseNet-121 generalized the best; we posit this was due to its heavy use of residual connections utilizing concatenation, which causes feature maps from earlier layers to be used deeper in the network, while aiding in gradient flow and regularization.
Keywords: Large Vessel Occlusion, Convolutional Neural Network, Deep Learning, Computed Tomography Angiography (CTA), Image Classification
Introduction
Large vessel occlusion (LVO) results in 90% of deaths among patients with acute ischemic stroke (AIS), while only occurring in a third of AIS cases1. Endovascular thrombectomy is effective for treating LVO, with LVO being resistant to thrombolytic drugs, an alternative form of treatment2. However, endovascular thrombectomy is not ubiquitously available, and requires transfer of patients to Level One Stroke Centers. Time is critical with AIS, with quicker treatment leading to better patient outcomes. Moreover, there exists a time window of 24 hours in which endovascular thrombectomy can be performed2, thus rapid identification of patients with LVO is imperative.
Tools from the field of artificial intelligence may facilitate the expedient identification of LVO3. CNNs have become the de facto standard for developing image learning approaches. Yet, the design and implementation of CNNs does not follow a single framework; CNN architectures can be composed from a pool of well-known and effective building blocks and connection techniques. Better understanding of how different architectural elements influence CNN performance for LVO detection on medical images could aid in designing improved CNNs for faster LVO detection and improved patient outcomes.
To provide context, the CNN Inception architecture saw heavy use after being introduced in 2014 and achieving state of the art performance with its use of the Inception module, a block of parallel processing branches joined through feature concatenation4. The following year, another CNN named ResNet came onto the scene, winning the ILSVRC classification task5. Designed as a response to the degradation problem, in which neural networks eventually decrease in performance as they get deeper, ResNet made use of skip / residual connections which combine residual information through addition, alleviating the degradation problem5. Shortly after, DenseNet was introduced and showed that high frequency of skip connections utilizing concatenation, rather than addition, to recombine residual information allowed for features produced by earlier layers to be reconsidered deeper in the network6. This innovation improved gradient flow and helped with regularization, making learning on small datasets easier and overfitting less likely6. As parameter-efficient neural networks for mobile devices became increasingly desired, as well as effective techniques for scaling models, EfficientNet-B0 was published as a baseline efficient CNN and was used to demonstrate the effectiveness of compound scaling7. CNNs are often adapted for domain specific applications; our model overview concludes with PhiNet, a small CNN designed to classify MRI contrasts8.
Artificial intelligence has been applied to LVO detection in several studies. A 3D Siamese CNN named DeepSymNet compared hemispheres of the brain and made use of a series of Inception modules to perform binary classification of LVO and determine size of ischemic cores in computed tomography angiography (CTA)9, 10. Other work showed binary classification of LVO was possible on multiphase CTA, utilizing a pretrained DenseNet-121, which had better initial results than ResNet-50, InceptionV3, and other CNNs: Xception and VGG16 on a dataset of 540 images11. In the realm of commercial software, RAPID for LVO detection was shown to be effective on CTA in single center12 and multicenter studies13. Other commercial software from Viz.ai makes use of a CNN to identify LVO on CTA14, 15. Use in clinical settings has shown that Viz.ai software can improve triage of patients with LVO, including increasing speed of patient transfer16, 17. With respect to preprocessing, maximum intensity projections (MIPs) have been used to facilitate detection of emergent LVO in CTA18, 19 with one study selecting a pre-trained and adapted ResNet-50 from a grouping of other pre-trained networks: InceptionV3, DenseNet-121 and NASNet for use on a dataset of over 1200 images19.
Neural networks have also been shown useful for improving CT image quality. Low-dose CT uses a smaller X-ray radiation dose, but results in noisier scans than traditional CT. With generative adversarial network (GAN) methods, noise can be reduced while maintaining important structural features20. Additionally, GAN super-resolution can be used to reconstruct higher resolution CT from lower resolution CT scans21. For medical image segmentation algorithms, unsupervised methods have been developed to handle domain adaptation between CT and magnetic resonance (MR) images when using a multi-modality approach22, as well as, purely on MR images, modern segmentation with contrastive learning utilizing a voxel-wise technique23.
Despite previous work using CNNs in LVO detection, as well as comparisons of models in specific medical imaging domain tasks 24, 25, 26, to the best of our knowledge there have been no studies explicitly examining how CNN architectural features affect performance of LVO binary classification on CTA. We are solely concerned with a detailed model comparison for our classification task of interest and on CTA data, rather than general image classification, or other medical imaging domain-specific classification tasks. In this work, we propose an analysis of various contemporary CNN architectures: ResNet-50, DenseNet-121, EfficientNet-B0, the current PhiNet, and a small CNN built around a single Inception module. We examine the performance differences between these models on our small dataset with a focus on how the number of learnable parameters and the type / frequency of skip connections affect model performance.
Materials and Methods
Data
Our dataset comprises 300 CTA scans of stroke-alerted patients at Vanderbilt University Medical Center between 2017 and 2019. These data were acquired in de-identified form under Institutional Review Board approval. The data are balanced with 50% of the scans depicting anterior circulation LVO, with all scans being selected from 677 stroke-alerted patients. In this paper LVO will be referring to occlusion in three locations each on both right and left sides, for a total of six artery groups. We include the first segment of the middle cerebral artery (M1), the proximal second segment of the middle cerebral artery (M2), and the internal carotid artery (ICA). The LVO positive scans are from subjects with the following characteristics: 56% male, 44% female; median age 65, interquartile range 55 to 76; 13% Black, 82% White, and 5% Other/Unknown. The LVO negative scans come from: 47% male, 53% female; with median age 66 and interquartile range 54 to 76; 17% Black, 79% White, and 3% Other/Unknown. Preprocessing of the scans began with head cropping and registration to a non-contrast computed tomography scan (NCCT) selected as template from our data. Additionally, a skull removal technique was applied to every NCCT and each mask was used on its corresponding CTA27. Subsequent manual Hounsfield unit window width and level adjustments were performed followed by the creation of 40 mm axial MIPs to optimally depict anterior circulation.
Models
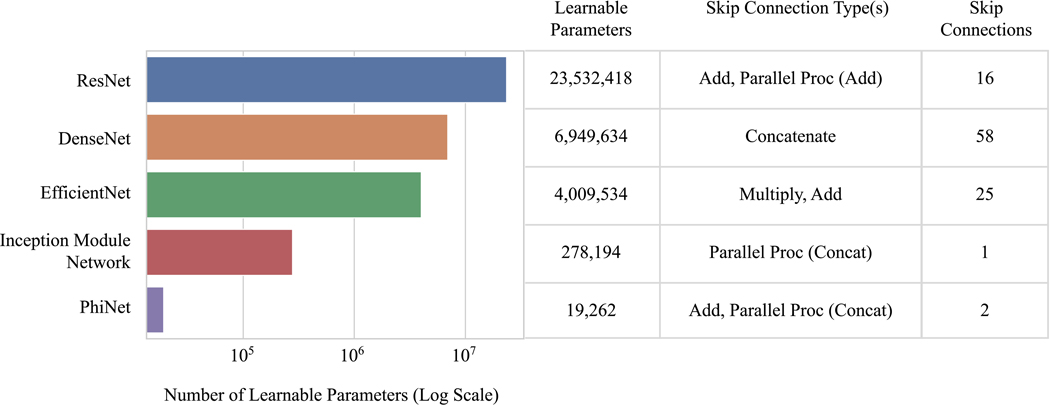
We compare five CNN architectures: ResNet-50, DenseNet-121, EfficientNet-B0, a current PhiNet, and an Inception module-based network, which consists of the first four layers of the Inception network4, fed into an Inception module, followed by global average pooling and a fully-connected layer. In terms of kernel size, blocks, etc., the architectures were left as default implementations, or in the case of the Inception module-based network, the utilized layers were from a default implementation. Note that the PhiNet architecture used is the current iteration from the model designer’s public code repository28. We focus on evaluating the performance of these models with respect to the number of learnable parameters and types and amount of skip connections (Figure 1).
Figure 1:
A comparison of the number of learnable parameters and the types and number of skip connections in our five models. Note that the skip connections type column lists operations which show how residual information is reintegrated when the skip / residual connection or parallel processing branch rejoins the network.
Cross-Validation & Threshold Moving
This procedure was identical for every model. The models were trained on a computer running Ubuntu 18.04.5 LTS and equipped with an NVIDIA GeForce GTX 1080 Ti graphics card. We performed 10-fold cross-validation over 240 of our 300 images (60 were withheld for external validation). Within the cross-validation, the splits for each fold specified 172 images for training, 44 images for validation, and 24 images for test. The splits did not specify balanced classes. To promote reproducibility, the images and splits seen by each model for a given fold were the same. The models trained on the entire MIP for each image, without using patches. Additionally, all models were trained with the same set of hyperparameters which were empirically chosen from an initial run of PhiNet. Each model used Adam as the optimizer, a learning rate of 1×10−4, batch size of eight, and was trained for at most 1000 epochs. Early stopping was used as regularization and was set to trigger after 200 epochs in which the validation accuracy did not improve by more than 1×10−8, which we use as the definition of convergence in this work. The model weights from the epoch with the best validation accuracy during training were selected for evaluation. To determine a suitable threshold for each model, the train, validation and test set predictions were aggregated over the 10 folds from cross-validation. Thresholded metrics (accuracy, precision, recall, specificity, and F1 score) were computed at every threshold from zero to one in increments of 1×10−3. The best threshold was selected where the standard deviation of the five metrics was the lowest.
Ablation Study
We proceeded to perform an ablation study on the best performing model on the cross-validation test sets. Every ablated model underwent identical training and threshold optimization as previously described. This study is explained further in the results section.
External Validation & Calibration
Each of the ten instances for each model were run on our balanced and withheld external validation set of 60 images. We call predictions from our trained models uncalibrated. To better calibrate the uncalibrated predictions, we use sigmoids of the form:
Here pi is the calibrated prediction, xi is a model instance’s uncalibrated prediction for an image and the parameters a and b are learned through gradient descent. Each instance of every model from cross-validation had its own sigmoid fit using its validation set predictions from cross-validation as input with regularization tuned on the cross-validation test predictions. The ith instance of each model then had its uniquely fit sigmoid used to calibrate its uncalibrated predictions on the withheld external validation set.
Results
Ablation Study
PhiNet was found to perform best on each of the five metrics for the cross-validation test sets. The base architecture consists of three main parallel processing branches. We examined six PhiNets in which we removed these main branches: three iterations consisted of each main branch isolated and three consisted of a combination of two of these main branches. Of these modifications, we found that a two-branch PhiNet containing the activated convolutional branch and pooling branch had better performance. Slight ablations were continuously performed on these branches. In total, 20 ablated models were created. The ablated PhiNet with best cross-validated performance is considered in following results.
Training and Detection Time
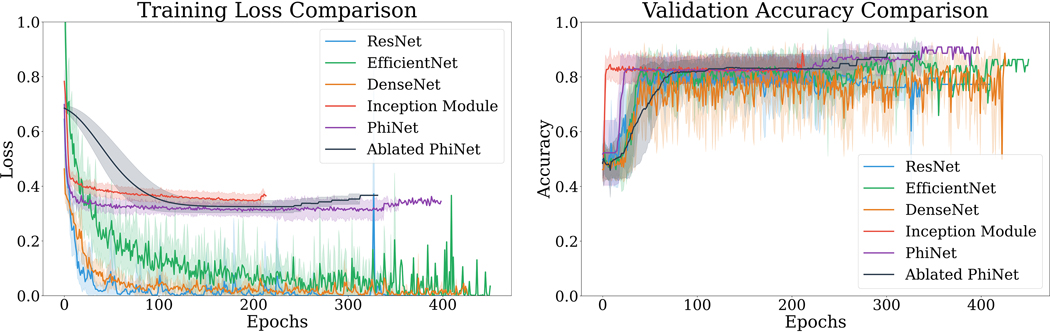
As previously stated, each cross-validation fold was split into a train, validation, and test partition. A comparison of training loss and validation accuracy during cross-validation is shown in Figure 2. Validation accuracy during training was used as criteria for convergence. Additionally, a comparison of training time and detecting time can be seen in Table 1.
Figure 2:
Mean training loss (left) and mean validation accuracy (right) for each fold are shown. Shaded regions represent ± the standard deviation.
Table 1.
The training time column lists the average seconds/step over 15 epochs, with batch size of eight. The detecting time column denotes the seconds/step for predicting on 60 images, with prediction batch size of one. The total epochs column gives the total epochs of training over the 10 folds from cross-validation.
| Training Time: Average Seconds / Step | Detecting Time | Total Epochs | |
|---|---|---|---|
|
ResNet
|
5.372 seconds | 1.015 seconds | 2808 |
|
DenseNet
|
6.539 seconds | 1.020 seconds | 3315 |
|
EfficientNet
|
6.413 seconds | 1.014 seconds | 3137 |
|
Inception Module
Network
|
1.037 seconds | 0.004 seconds | 2080 |
|
PhiNet
|
1.039 seconds | 0.005 seconds | 2652 |
| Ablated PhiNet | 0.087 seconds | 0.004 seconds | 2748 |
Ensemble Predictions by Artery Group
We look to results on the withheld external validation set to examine the performance of the models on LVO-positive images from six main artery groups and on LVO-negative images. Figure 3 shows accuracy of ensemble predictions using majority voting for each group. Using this ensemble prediction method, DenseNet-121 and EfficientNet-B0 perform equally and best on the withheld external validation set.
Figure 3:
An ensemble of the ten instances of each model was used to generate binary class decisions through majority voting for every image in the withheld external validation set. Each table entry shows prediction accuracy over the images in a given artery group for a model’s ensemble. Artery groups and the LVO-negative group are given at the top of the columns along with the number of images for the group, and an example MIP. The Inception module-based network is abbreviated to Inc-Mod and PhiNet is abbreviated to Phi.
Mean Metrics and Calibration
To look more closely at average model performance, we consider mean metrics instead of ensemble predictions. These results are shown over the cross-validation test sets, the withheld external validation set, and the withheld external validation set with calibrated predictions (Table 2). Further, we wish to know if there are any significant differences among the performances for our group of models. For each model and on a given metric, the performance on every fold was aggregated into a vector for that model on that metric. This was done separately for the set of metrics: accuracy, precision, recall, specificity, and F1 score. The Friedman test was performed over the collection of vectors for every model for a given metric. This process was performed separately for the cross-validation test, withheld external validation, and calibrated withheld external validation sets. The Bonferroni correction was applied and statistically significant metric columns are denoted in Table 2. Additionally, assessment of model calibration was performed using the integrated calibration index (ICI), a weighted numeric calibration metric that considers prediction distribution29. The ICI is reported on the external validation set predictions before and after the sigmoid calibration method was applied (Table 2).
Table 2.
Accuracy (ACC), precision (PR), recall (RE), specificity (SP), and F1 Score (F1) are reported for each of the five original models and the best-ablated PhiNet. The 10-fold cross-validation created 10 trained instances of each model–the metrics reported here are mean metrics computed over each model’s 10 instances’ performances ± the standard deviation. “Cross-validation test” refers to the test portion of each fold. There are 24 distinct images in each of the 10 cross-validation test folds. 60 images are in the withheld external validation set, which each of the 10 instances of every model saw. Statistically significant columns are denoted with *, where the Bonferroni corrected p-value < 0.05 for the Friedman test for the group of metric vectors from which the column is derived. Additionally, the ICI is reported to show how well-calibrated each model is on the withheld external validation set, before and after the sigmoid calibration method is applied. Bolded values represent the best performance per column for the first five models. The best-ablated PhiNet is separated from the original five models due to its further architecture optimization. In the cross-validation test results we can see that the PhiNet ablation has bolded metrics, this is due to its performance surpassing that of the other five models.
| Cross-Validation Test | External Validation | Calibrated External Validation | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||||
| ACC | PR | RE | SP | F1 | ACC | PR* | RE | SP | F1 | ICI | ACC* | PR* | RE | SP* | F1 | ICI | |
|
ResNet
|
0.779 ± 0.098 | 0.782 ± 0.125 | 0.820 ± 0.195 | 0.770 ± 0.139 | 0.778 ± 0.108 | 0.772 ± 0.045 | 0.733 ± 0.036 | 0.857 ± 0.092 | 0.687 ± 0.055 | 0.788 ± 0.049 | 0.145 | 0.752 ± 0.103 | 0.664 ± 0.237 | 0.780 ± 0.289 | 0.723 ± 0.112 | 0.716 ± 0.257 | 0.096 |
|
DenseNet
|
0.800 ± 0.098 | 0.810 ± 0.133 | 0.802 ± 0.172 | 0.822 ± 0.125 | 0.791 ± 0.115 | 0.830 ± 0.029 | 0.792 ± 0.020 | 0.897 ± 0.074 | 0.763 ± 0.037 | 0.839 ± 0.036 | 0.103 | 0.838 ± 0.025 | 0.796 ± 0.021 | 0.910 ± 0.039 | 0.767 ± 0.027 | 0.849 ± 0.025 | 0.073 |
|
EfficientNet
|
0.812 ± 0.088 | 0.824 ± 0.147 | 0.816 ± 0.133 | 0.829 ± 0.141 | 0.808 ± 0.096 | 0.803 ± 0.034 | 0.767 ± 0.026 | 0.873 ± 0.084 | 0.733 ± 0.047 | 0.815 ± 0.040 | 0.130 | 0.768 ± 0.099 | 0.688 ± 0.242 | 0.777 ± 0.283 | 0.760 ± 0.091 | 0.728 ± 0.258 | 0.087 |
|
Inception Module
Network
|
0.825 ± 0.087 | 0.846 ± 0.133 | 0.832 ± 0.233 | 0.838 ± 0.144 | 0.805 ± 0.150 | 0.783 ± 0.100 | 0.752 ± 0.061 | 0.823 ± 0.280 | 0.743 ± 0.097 | 0.759 ± 0.213 | 0.104 | 0.825 ± 0.018 | 0.770 ± 0.012 | 0.927 ± 0.041 | 0.723 ± 0.022 | 0.841 ± 0.019 | 0.087 |
| PhiNet | 0.850 ± 0.081 | 0.858 ± 0.129 | 0.871 ± 0.141 | 0.852 ± 0.152 | 0.850 ± 0.083 | 0.793 ± 0.034 | 0.776 ± 0.026 | 0.830 ± 0.117 | 0.757 ± 0.063 | 0.797 ± 0.050 | 0.119 | 0.822 ± 0.016 | 0.769 ± 0.016 | 0.920 ± 0.032 | 0.723 ± 0.032 | 0.838 ± 0.015 | 0.092 |
|
| |||||||||||||||||
| Ablated PhiNet | 0.862 ± 0.074 | 0.873 ± 0.119 | 0.871 ± 0.113 | 0.877 ± 0.118 | 0.862 ± 0.072 | 0.798 ± 0.018 | 0.776 ± 0.015 | 0.840 ± 0.058 | 0.757 ± 0.032 | 0.806 ± 0.026 | 0.144 | 0.825 ± 0.018 | 0.774 ± 0.007 | 0.917 ± 0.036 | 0.733 ± 0.000 | 0.839 ± 0.019 | 0.105 |
Discussion
Cross-Validation Test Performance: Five Starting Models
PhiNet outperformed every starting model on accuracy, precision, recall, specificity, and F1 score (Table 2). We noticed a general trend over these thresholded metrics that corresponds to the number of learnable parameters in these models. Fewer learnable parameters in a model led to better average performance on the cross-validation test sets. This trend exactly applies to accuracy, precision, and specificity where model performance ranking from best to worst directly corresponds to sorting the models from smallest to largest. For recall and F1 score the trend approximately applies—there are slight exceptions with ResNet-50 performing better on recall and EfficientNet-B0 performing better on the F1 score than the general trend.
Cross-Validation Test Performance: Ablated PhiNet
When the cross-validation test performance of the best-ablated PhiNet is included, this trend of smaller models performing better holds (Table 2). This ablated PhiNet has notably fewer learnable parameters than PhiNet, with only 475 compared to PhiNet’s 19,262. The ablated PhiNet performs better than all the other models across accuracy, precision, recall, specificity and F1 score.
Training & Detection
When doing a rough split of the models into two groups: smaller (Inception module-based network, PhiNet, best-ablated PhiNet) and larger (ResNet-50, DenseNet-121, EfficientNet-B0), we can see in Figure 2 that the smaller models were not able to achieve as great of a reduction in training loss as the larger models. Additionally, we can see in Table 1 that the smaller models were much faster for training and detection in terms of seconds/step, when compared with the larger models.
External Validation Performance and External Validation Calibration
For the ensemble predictions, DenseNet-121 and EfficientNet-B0 both perform equally and best. However, when solely looking at binary classification of LVO and choosing mean performance of the ten instances of each model instead of ensemble predictions by majority voting, we can extract more detailed performance information. The pattern of smaller models performing better disappears on the withheld external validation set. DenseNet-121, the model with the second largest number of learnable parameters comes out as definitively the strongest model, performing better than all other models, including EfficientNet-B0 and the ablated PhiNet, on the withheld external validation set. This implies that DenseNet-121 had superior generalization.
The success of DenseNet-121 is likely due to its heavy use of skip connections that combine residual information using concatenation. While other models in our grouping utilize concatenation to combine parallel processing branches, DenseNet-121 is the only model to use concatenation to rejoin skip connections that directly connect previous layer outputs as extra inputs deeper in the network. Additionally, DenseNet-121 uses notably more skip connections than any of our other models (Figure 1). After calibration of the external validation predictions, DenseNet-121 still performs best on all metrics except for recall. Additionally, DenseNet-121 was the best-calibrated model both before and after the sigmoid calibration methods, based on assessment by ICI. An important note for DenseNet-121’s performance is that our dataset is small (n=300), and the effectiveness of its skip connections may not hold true for larger datasets. In the original paper on DenseNet, it was commented that this type of skip connection benefits performance on smaller datasets6, which we find to be true on our small dataset.
Discussion of Model Differences
From our results, it is clear that with fixed LVO data, hyperparameters, and experiments, different CNN architectures have diverse outcomes with regards to training time, evaluation time, training loss convergence, calibration, performance, generalization, and other measurable outcomes. When selecting a CNN architecture for an LVO detection task, careful consideration of tradeoffs, including dataset size, training time, and evaluation time, should be made centered around the specific application for which a model will be employed.
Conclusions
While models with fewer learnable parameters performed better during cross-validation, this pattern did not hold when checking performance on our external validation set. Note that the external validation set was not revealed during any model design, evaluation, or tuning. We applied the frozen models from cross-validation only once to the external validation set and no new model training was performed after the external validation set was revealed. We conclude that of the examined models, DenseNet-121 generalized the best to our small dataset of LVO MIPs based on its superior performance on the withheld external validation set. It appears that this is due to its heavy use of skip connections that recombine residual information using concatenation which increases consideration of feature maps from earlier in the network, while also improving gradient flow and regularization. This architectural feature may prove useful when selecting models for optimized performance on LVO detection when training on small datasets, as well as for considering adaptations to existing model architectures and constructing novel architectures in this domain. The differences between performance in cross-validation and external validation highlight the importance of single pass study designs and the possibility of information contamination during model design / ablation.
Acknowledgements
We would like to thank Kiersten Espaillat, Cailey Kerley, Karthik Ramadass, and Praitayini Kanakaraj. This material is partially supported by the National Science Foundation Graduate Research Fellowship under Grant No. DGE-1746891 as well as ViSE/VICTR VR3029. The image dataset was obtained in part from ImageVU, a research resource supported by the VICTR CTSA award (ULTR000445 from NCATS/NIH), Vanderbilt University Medical Center institutional funding and Patient-Centered Outcomes Research Institute (PCORI; contract CDRN-1306-04869). We extend gratitude to NVIDIA for their support by means of the NVIDIA hardware grant. This work is supported by NIH grant R01GM120484. Funds also came from NSF 1452485 (Landman). This work was conducted in part using the resources of the Advanced Computing Center for Research and Education at Vanderbilt University, Nashville, TN. The project described was supported by the National Center for Research Resources, Grant 1UL1RR024975-01, and is now at the National Center for Advancing Translational Sciences, Grant 2UL1TR000445-06.
Footnotes
Data Availability Statement
Research data are not shared.
Conflicts of Interest
The authors have no relevant conflicts of interest to disclose.
References
- 1.Malhotra K, Gornbein J, Saver JL. Ischemic strokes due to large-vessel occlusions contribute disproportionately to stroke-related dependence and death: A review. Front Neurol. 2017;8(NOV). doi: 10.3389/fneur.2017.00651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rennert RC, Wali AR, Steinberg JA, et al. Epidemiology, Natural History, and Clinical Presentation of Large Vessel Ischemic Stroke. Clin Neurosurg. 2019;85. doi: 10.1093/neuros/nyz042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamal H, Lopez V, Sheth SA. Machine learning in acute ischemic stroke neuroimaging. Front Neurol. 2018;9(NOV). doi: 10.3389/fneur.2018.00945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Szegedy C, Liu W, Jia Y, et al. Going deeper with convolutions. In: Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition. Vol 07–12-June-2015.; 2015. doi: 10.1109/CVPR.2015.7298594 [DOI] [Google Scholar]
- 5.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition. Vol 2016-December.; 2016. doi: 10.1109/CVPR.2016.90 [DOI] [Google Scholar]
- 6.Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely connected convolutional networks. In: Proceedings - 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017. Vol 2017-Janua.; 2017. doi: 10.1109/CVPR.2017.243 [DOI] [Google Scholar]
- 7.Tan M, Le Q V. EfficientNet: Rethinking model scaling for convolutional neural networks. In: 36th International Conference on Machine Learning, ICML 2019. Vol 2019-June.; 2019. [Google Scholar]
- 8.Remedios S, Pham DL, Butman JA, Roy S. Classifying magnetic resonance image modalities with convolutional neural networks. In: Proc. SPIE 10575, Medical Imaging 2018: Computer-Aided Diagnosis.; 2018. doi: 10.1117/12.2293943 [DOI] [Google Scholar]
- 9.Barman A, Inam ME, Lee S, Savitz S, Sheth S, Giancardo L. Determining ischemic stroke from ct-angiography imaging using symmetry-sensitive convolutional networks. In: Proceedings - International Symposium on Biomedical Imaging. Vol 2019-April.; 2019. doi: 10.1109/ISBI.2019.8759475 [DOI] [Google Scholar]
- 10.Sheth SA, Lopez-Rivera V, Barman A, et al. Machine Learning-Enabled Automated Determination of Acute Ischemic Core From Computed Tomography Angiography. Stroke. 2019;50(11). doi: 10.1161/STROKEAHA.119.026189 [DOI] [PubMed] [Google Scholar]
- 11.Stib MT, Vasquez J, Dong MP, et al. Detecting large vessel occlusion at multiphase CT angiography by using a deep convolutional neural network. Radiology. 2020;297(3). doi: 10.1148/radiol.2020200334 [DOI] [PubMed] [Google Scholar]
- 12.Amukotuwa SA, Straka M, Smith H, et al. Automated detection of intracranial large vessel occlusions on computed tomography angiography a single center experience. Stroke. 2019;50(10). doi: 10.1161/STROKEAHA.119.026259 [DOI] [PubMed] [Google Scholar]
- 13.Amukotuwa SA, Straka M, Dehkharghani S, Bammer R. Fast automatic detection of large vessel occlusions on CT angiography. Stroke. 2019;50(12). doi: 10.1161/STROKEAHA.119.027076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barreira C, Bouslama M, Lim J, et al. E-108 Aladin study: automated large artery occlusion detection in stroke imaging study – a multicenter analysis. In:; 2018. doi: 10.1136/neurintsurg-2018-snis.184 [DOI] [Google Scholar]
- 15.Chatterjee A, Somayaji NR, Kabakis IM. Abstract WMP16: Artificial Intelligence Detection of Cerebrovascular Large Vessel Occlusion - Nine Month, 650 Patient Evaluation of the Diagnostic Accuracy and Performance of the Viz.ai LVO Algorithm. In: Stroke. Vol 50.; 2019. doi: 10.1161/str.50.suppl_1.wmp16 [DOI] [Google Scholar]
- 16.Morey JR, Fiano E, Yaeger KA, Zhang X, Fifi JT. Impact of Viz LVO on Time-to-Treatment and Clinical Outcomes in Large Vessel Occlusion Stroke Patients Presenting to Primary Stroke Centers. [published online July 5, 2020]. doi: 10.1101/2020.07.02.20143834 [DOI] [Google Scholar]
- 17.Hassan AE, Ringheanu VM, Rabah RR, Preston L, Tekle WG, Qureshi AI. Early experience utilizing artificial intelligence shows significant reduction in transfer times and length of stay in a hub and spoke model. Interv Neuroradiol. 2020;26(5). doi: 10.1177/1591019920953055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stib MT, Dong MP, Kim YH, et al. Deep Learning in Emergent Large Vessel Occlusion Detection using Maximum Intensity Projections via CT Angiography. 2018;(March):1–3. [Google Scholar]
- 19.Dong M, Kim A, Subzwari S, et al. Deep Learning for ELVO Stroke Detection. In: Machine Intelligence Conference 2018.; 2018. [Google Scholar]
- 20.You C, Yang Q, Shan H, et al. Structurally-Sensitive Multi-Scale Deep Neural Network for Low-Dose CT Denoising. IEEE Access. 2018;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.You C, Li G, Zhang Y, et al. CT Super-Resolution GAN Constrained by the Identical, Residual, and Cycle Learning Ensemble (GAN-CIRCLE). IEEE Trans Med Imaging. 2020;39(1). [DOI] [PubMed] [Google Scholar]
- 22.You C, Yang J, Chapiro J, Duncan JS. Unsupervised Wasserstein Distance Guided Domain Adaptation for 3D Multi-domain Liver Segmentation. In: Interpretable and Annotation-Efficient Learning for Medical Image Computing.; 2020:155–163. [Google Scholar]
- 23.You C, Zhao R, Staib L, Duncan JS. Momentum Contrastive Voxel-wise Representation Learning for Semi-supervised Volumetric Medical Image Segmentation. Published online 2021:1–11. http://arxiv.org/abs/2105.07059 [DOI] [PMC free article] [PubMed]
- 24.Setio AAA, Traverso A, de Bel T, et al. Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: The LUNA16 challenge. Med Image Anal. 2017;42:1–13. doi: 10.1016/j.media.2017.06.015 [DOI] [PubMed] [Google Scholar]
- 25.Baltruschat IM, Nickisch H, Grass M, Knopp T, Saalbach A. Comparison of Deep Learning Approaches for Multi-Label Chest X-Ray Classification. Sci Rep. 2019;9(1). doi: 10.1038/s41598-019-42294-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang Z, Zhu X, Ding M, Zhang X. Medical Image Classification Using a Light-Weighted Hybrid Neural Network Based on PCANet and DenseNet. IEEE Access. 2020;8. https://ieeexplore.ieee.org/stamp/stamp.jsp?arnumber=8979430 [Google Scholar]
- 27.Muschelli J, Ullman NL, Mould AW, Vespa P, Hanley DF, Crainiceanu CM. Validated Automatic Brain Extraction of Head CT Images. Neuroimage. 2015;114:379–385. doi: 10.1016/j.neuroimage.2015.03.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Remedios SW. PhiNet. https://github.com/sremedios/phinet. Accessed September 1, 2020.
- 29.Austin PC, Steyerberg EW. The Integrated Calibration Index (ICI) and Related Metrics for Quantifying the Calibration of Logistic Regression Models. Stat Med. 2019;38(21):4051–4065. doi: 10.1002/sim.8281 [DOI] [PMC free article] [PubMed] [Google Scholar]