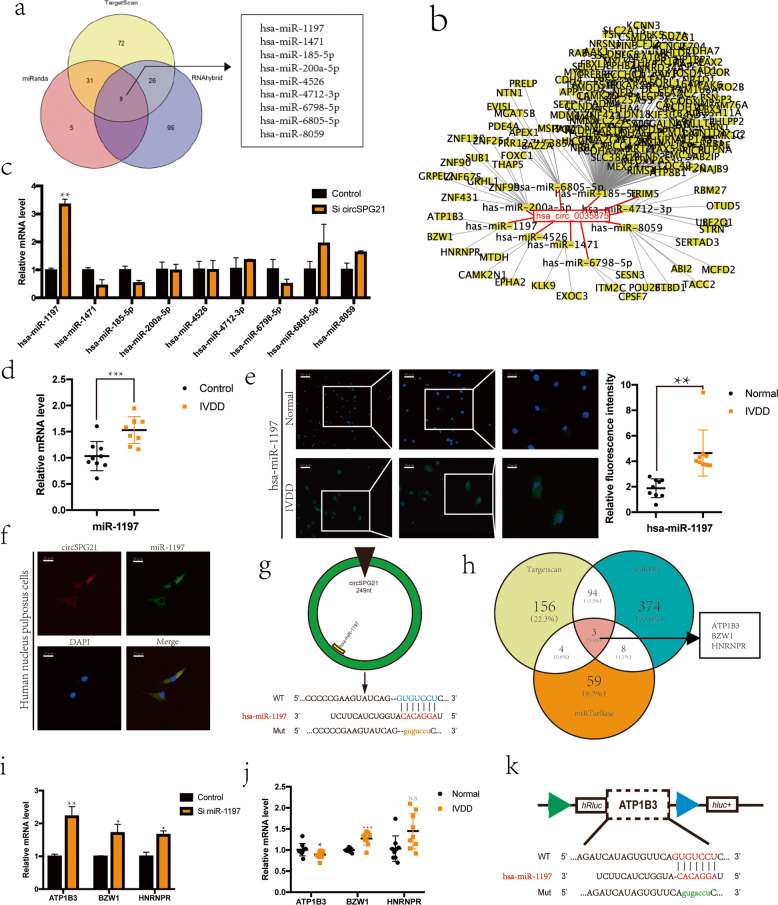
Fig. 3. ceRNA network construction.
a circSPG21-target microRNA was predicted based on the intersection of the TargetScan, miRanda, and RNAhybrid software data. b A ceRNA network diagram was drawn using Cytoscape. c Expression of microRNAs after circSPG21 knockdown, as detected by real-time quantitative polymerase chain reaction (RT-qPCR), **p < 0.01. Data are presented as the mean ± S.D., and the p values were determined by a two-tailed unpaired Student’s t-test. d miR-1197 expression increased in degenerated tissues, as measured by RT-qPCR; n = 6, ***p < 0.001. Data are presented as the mean ± S.D., and the p values were determined by a two-tailed unpaired Student’s t-test. e Left, the expression of miR-1197, as measured by fluorescence in situ hybridization (FISH), in degenerated tissues was higher than that in control tissues (scale bar, 100 μm). Right panel, the fluorescence intensity of miR-1197 was stronger in degenerated tissue (two different donors), **p < 0.01. Data are presented as the mean ± S.D., and the p values were determined by a two-tailed unpaired Student’s t-test. The miR-1197 probe was labeled with Alexa Fluor 488. f circSPG21 and miR-1197 were localized in the cell, and both were seen in the cytoplasm (scale bar, 25 μm). g The sites where circSPG21 and miR-1197 bind to each other were identified using CircInteractome. h The target genes of miR-1197 were predicted based on the intersection of TargetScan, miRDB, and miRTarBase results. i Expression of target genes after miR-1197 knockdown, as detected by RT-qPCR; *p < 0.05, **p < 0.01. Data are presented as the mean ± S.D., and the p values were determined by a two-tailed unpaired Student’s t-test. j The expression of ATP1B3, BZW1, and HNRNPR was detected in degenerated tissues using RT-qPCR; *p < 0.05, ***p < 0.001. Data are presented as the mean ± S.D., and the p values were determined by a two-tailed unpaired Student’s t-test. k The binding site of miR-1197 and ATP1B3 predicted by TargetScan.