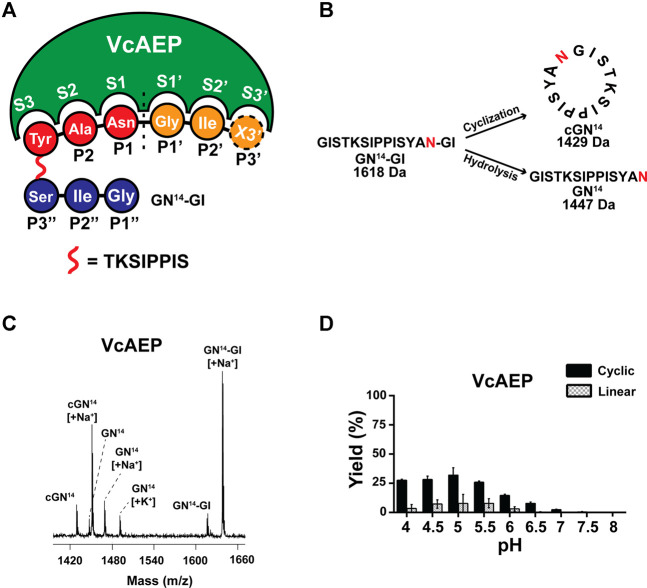
FIGURE 2.
Analysis of VcAEP AEP activity. (A) Nomenclature of AEP/PAL and substrate, using the peptide substrate GN14-GI (GISTKSIPPISYAN-GI) as a model. Based on the nomenclature of substrate binding of protease proposed by Schechter and Berger, the cleavage site of the substrate is named P1 (red), and the corresponding binding pocket of the enzyme is S1. After cleavage, the amino acid residues of leaving group are P1′, P2′, P3′ Etc (orange). For PALs to cyclize or ligate substrate, the amino acid residues of the incoming group are P1″, P2″, P3″ etc., (blue). X stands for the 20 amino acids. (B) Schematic representations of AEP-mediated cyclization and hydrolysis of the 16-residue peptide substrate GN14N-GI (MW 1618 Da). The P1-Asn position is colored red. The enzymatic reaction of VcAEP yields two products: the 14-residue cyclic product cGN14 (1429 Da) and 14-residue linear product GN14 (1447 Da) with release of a GI dipeptide. (C) Representative MALDI-TOF mass spectrometry of the cyclic product cGN14 and the linear product GN14 generated by VcAEP together with GN14N-GI as the starting material. The reaction was conducted at pH 6, at 37°C for 15 min with a 1:20 molar ratio of enzyme:substrate. (D) Product yields of VcAEP wild type were quantified by calculating the peak areas of MALDI-TOF mass spectrometry. Gray and black bars indicate the percentage yield of linear product and cyclic product, respectively. The reactions were conducted at 37°C for 15 min with a molar enzyme:substrate ratio of 1:20 (VcAEP). Average yield and SDs were calculated from experiments performed in triplicate.