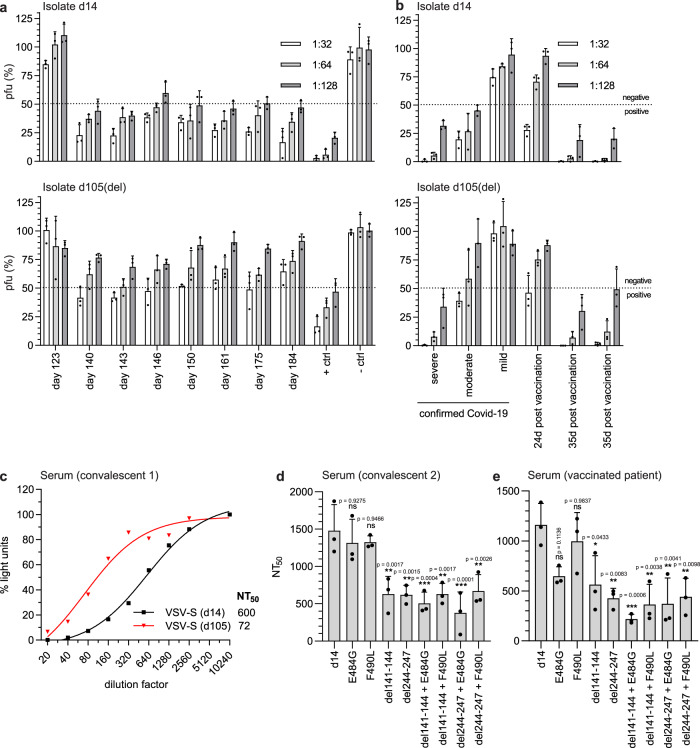
Fig. 5. Delayed seroconversion and viral escape from the spike protein-specific antibody response.
a, b Detection of neutralizing activity of immune sera against SARS-CoV-2 variants. 100 pfu of the d14 and d105(del) isolates were incubated for 60 min at room temperature with serial dilutions of the patient sera. Sera obtained from naïve (– ctrl) or convalescent individuals (+ ctrl) served as negative and positive controls. Virus neutralization was determined by plaque assay on VeroE6 cells. Virus titers are indicated as percentages (mean ± SD) of the titer of the untreated virus inoculum. The dotted lines indicate the cutoff value between positive (<50%) and negative (>50%) neutralization. Shown are the means of three biological replicates. a Sera from the immunocompromised patient. The times of blood withdrawal are indicated. b Convalescent sera from COVID-19 patients suffering from mild, moderate, or severe disease or human post-vaccination (BNT162b2 mRNA) sera. c–e Neutralization capacity of SARS-CoV-2 antisera using VSV*∆G(FLuc) vector pseudotyped with the SARS-CoV-2 spike protein and coding for firefly luciferase. The pseudotyped viruses were incubated with serial dilutions of a COVID-19 convalescent serum prior to inoculation of VeroE6 cells. Pseudotyped virus infection was monitored 16 h post-infection by measuring the firefly luciferase activity in the cell lysates. The control without serum was set to 100%. c Neutralization of VSV*∆G(FLuc) pseudotyped with the early and late SARS-CoV-2 spike variants (d14 and d105). d, e Neutralization of VSV*∆G(FLuc) pseudotyped with the d14 spike protein containing the individual or combined mutations found in the late d105 and d140 sequences. Immune sera from two different convalescent COVID-19 patients (c, d) or a vaccinated person (e) were analyzed. The neutralization was determined by calculating the NT50 via a non-linear regression (variable slope, four parameters). Shown are means ± SD (n = 3). Statistics were calculated with a one-way ANOVA (Tukey’s multiple comparison test), ns = non-significant, *p < 0.05, **p < 0.01, ***p < 0.001. The exact p-values are given in the figure. Source data are provided as a Source Data file.