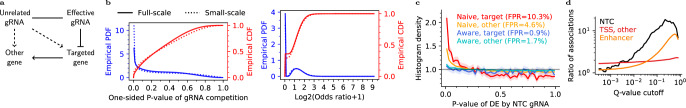
Figure 5. Normalisr reduced the false positives from gRNA cross-associations in high-MOI single-cell CRISPR screens.
a Example scenario of false positives of gRNA–gene associations (dashed) arising from negative gRNA cross-associations and true regulation (solid) for genes targeted directly by the effective gRNA or indirectly through the targeted genes (other gene). b Detection of different gRNAs were anti-correlated across cells in terms of one-sided hypergeometric P values (left, X) and odds ratios (right, X) between gRNA pairs. Empirical PDFs and CDFs are shown in blue (left Y) and red (right Y), respectively. c Ignoring untested gRNAs increased FPR, as shown by density histogram (Y) of CRISPRi DE two-sided P values from NTC gRNAs (X). P value histograms and FPRs were computed separately with competition-naive (ignoring untested gRNAs) and competition-aware (regarding untested gRNAs as covariates) methods and separately for genes targeted by positive control gRNAs at the TSS and other genes. Gray line indicates the expected uniform distribution for null P value. Shades indicate absolute errors estimated as , where N is the count in each bin. d Competition-naive DE inflated the number of significant gRNA–gene associations relative to the competition-aware method (Y) at different nominal Q-values (X).