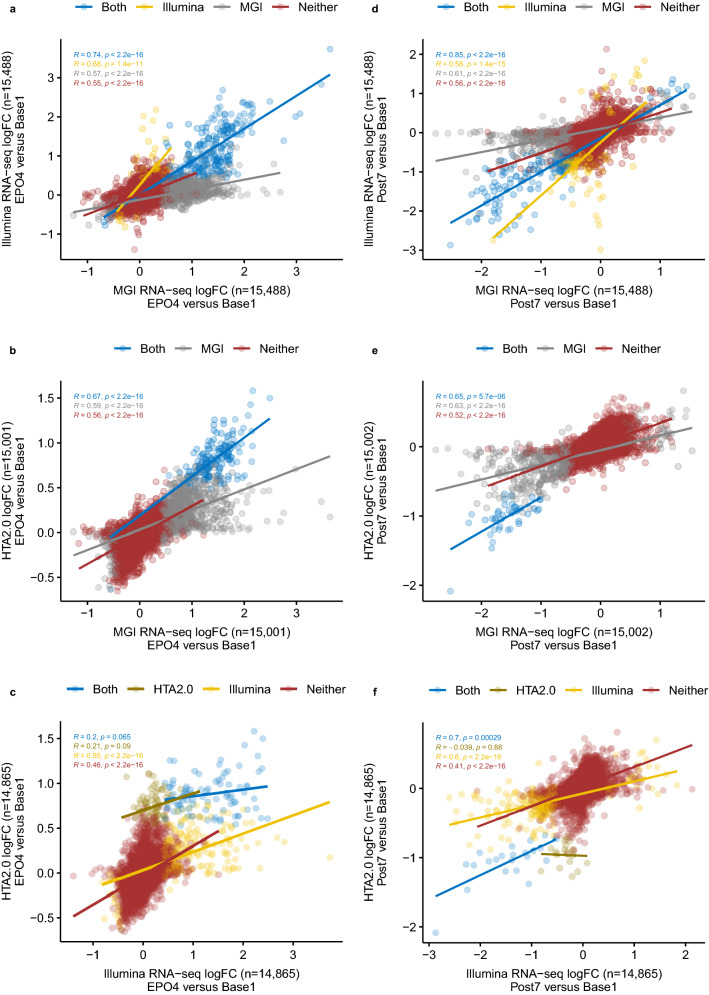
Figure 2.
Cross-platform gene expression correlation analyses of log2-transformed fold changes of all identified gene features. (a–c) Genes identified when compared the level of expression between EPO4 and Base1 among the platform pairs of Illumina-MGI RNA-seq (a), GeneChip HTA2.0-MGI RNA-seq (b), GeneChip HTA2.0-Illlumina RNA-seq (c). d-f Genes identified when compared the level of expression between Post7 and Base1 among the platform pairs of Illumina-MGI RNA-seq (d), GeneChip HTA2.0-MGI RNA-seq (e), GeneChip HTA2.0-Illlumina RNA-seq (f). Only protein-coding genes are included here for cross-platform comparisons. Genes identified as differentially expressed by each pair are plotted in blue; genes that are only differentially expressed in Illumina RNA-seq, MGI RNA-seq or GeneChip HTA2.0 are plotted in yellow, grey and dijon, respectively; genes not identified as differentially expressed by a pair are plotted in red. For simplicity, the maximum expression value of a gene was used when multiple transcript cluster IDs or Ensembl gene IDs were mapped to the same gene symbol. FOXO3B is only differentially expressed in GeneChip HTA2.0 when compared to the MGI RNA-seq findings in (b), thus it has been removed from the correlation analysis. R: Pearson’s r. LogFC: log2-transformed fold change.