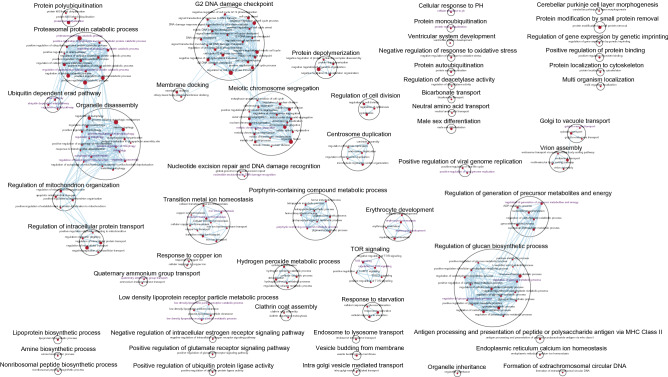
Figure 3.
Biological network of the MGI RNA-seq dataset following Gene Ontology (biological process) gene set enrichment analysis in GSEA (v4.0.3) and visualisation in Cytoscape (3.8.0). Each circle (node) represents a gene set and two nodes are connected by lines (edges) indicating shared genes. The size of a node and width of an edge are proportional to the number of genes enriched in a gene set and the number of genes shared between gene sets, respectively. Gene sets that are similar were annotated and clustered to form a biological theme using the AutoAnnotate App in Cytoscape. The most significantly enriched gene set is used to label a gene set cluster, defined by NES. Red node: gene set enriched in EPO4. Purple node label: top gene sets with NES > 1.90. The enrichment map was created with pathway FDR < 0.1, nominal P < 0.05 and Jaccard Overlap coefficient > 0.375 with combined constant k = 0.5.