Figure 2.
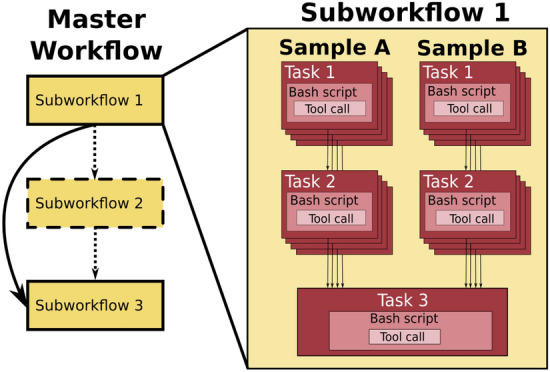
Bioinformatics workflows with multiple levels of complexity warrant a modular construction. It is easiest to program the workflow when its logic is abstracted away (in Tasks, red) from the command line invocations (in Bash scripts, pink) of the bioinformatics tools (light pink). Individual workflows can be further used as subworkflows of a larger Master workflow (e.g., Fig Supplementary 1). This architecture facilitates expression of additional complexity due to optional modules (dashed line), nested levels of parallelism (groups of arrows connecting red rectangles) and scatter-gather patterns (task 2 scattered across samples being merged into task 3).
