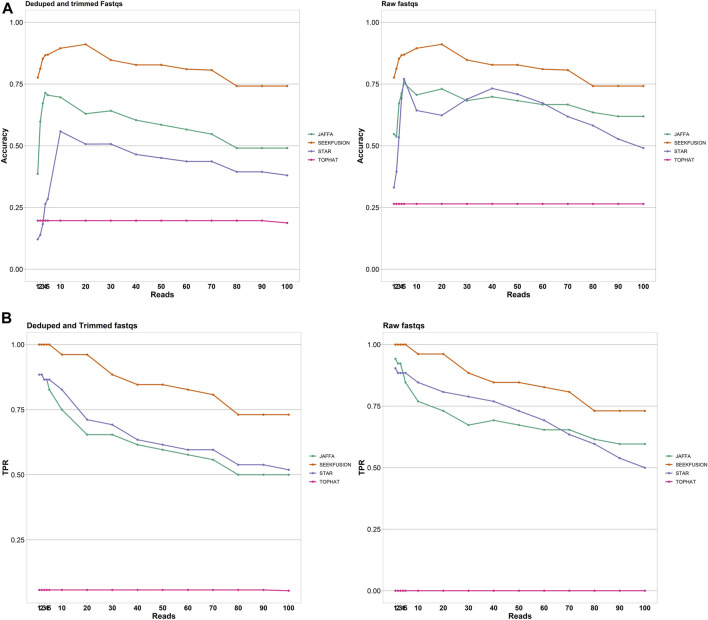
FIGURE 4.
(A) Accuracy of the tools was assessed for the tools JAFFA, STAR-Fusion, Tophat Fusion and SeekFusion at varying read support thresholds. SeekFusion demonstrated highest accuracy in detecting the gene fusions at all levels of read support. The median accuracy of SeekFusion was 82% followed by JAFFA-Hybrid at 60%, STAR-Fusion at 39% and Tophat-Fusion at 20% for Mode1. SeekFusion was the only pipeline that demonstrated high accuracy in detecting fusions from raw FASTQs. (B) True Positive Rate of the tools was assessed for the tools JAFFA, STAR-Fusion, Tophat Fusion and SeekFusion at varying read support thresholds. SeekFusion demonstrated the highest sensitivity in detecting the gene fusions at all levels of read support. For the deduped and trimmed FASTQs, we observed that the median true positive rate of SeekFusion was 89% followed by STAR-Fusion at 69% and JAFFA-Hybrid at 65%. Tophat Fusion performed least favorably among the tools assessed with a median true positive rate of 1%. SeekFusion was the only tool that successfully detected true positives in the raw FASTQs.