Figure 1.
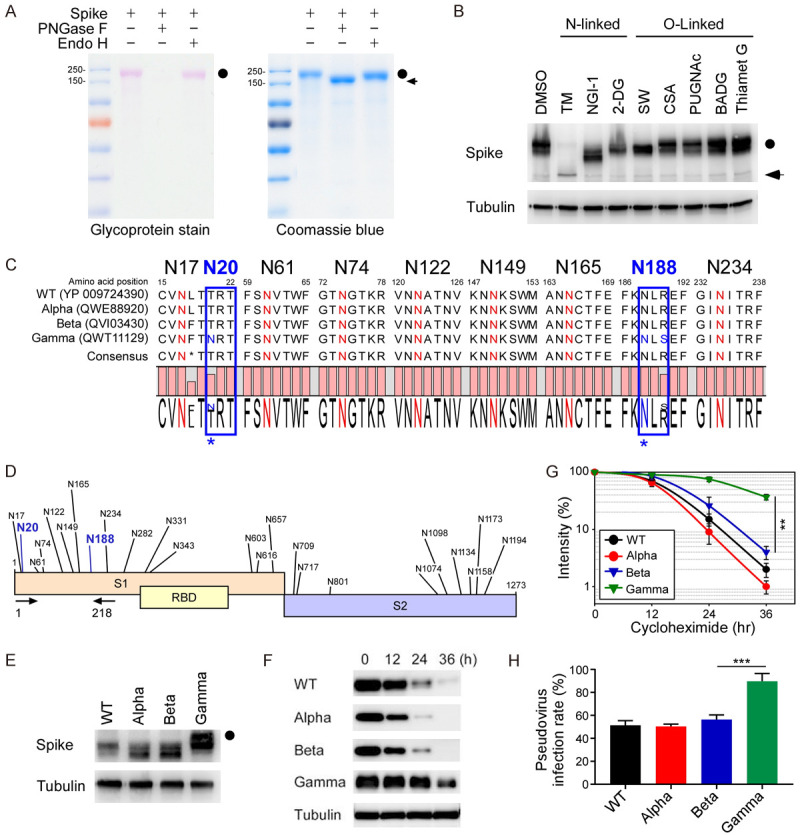
Hyperglycosylated S of SARS-CoV-2 Gamma variant is more stable in cells. A. The glycosylation pattern of the S protein. S protein was treated with PNGase F or Endo H and analyzed by glycoprotein staining (left panel) and Coomassie blue staining (right panel). Black circle, glycosylated S; arrowhead, non-glycosylated S. B. Western blot analysis of S protein expression pattern treated with N-linked and O-linked glycosyltransferase inhibitors. Circle, glycosylated S; arrow, non-glycosylated S. C. Amino acid sequences alignment of SARS-CoV-2 WT, Alpha, Beta, and Gamma variants for evolutionarily conserved NXT/S motifs. D. Diagram of NXT/S motifs in S protein of SARS-CoV-2. Conserved glycosylation sites on the S of Gamma variant are labeled in black. Two additional NTX motifs on the S of Gamma variant are marked in blue. Arrowhead indicates the PCR primers used to amplify the S of WT, Alpha, Beta, and Gamma variants. E. Western blot analysis of S protein (1-218 amino acids) of WT, Alpha, Beta, and Gamma variants. Circle, hyper-glycosylated Gamma variant. F. Western blot analysis of S protein expression in the cycloheximide-chase assay. G. The intensity of S protein in SARS-CoV-2 WT, Alpha, Beta, and Gamma variants was quantified using a densitometer. H. Luciferase activity was measured in HEK293T-ACE2 cells infected with SARS-CoV-2 pseudovirus generated from wild-type and mutant spike SARS-CoV-2.
