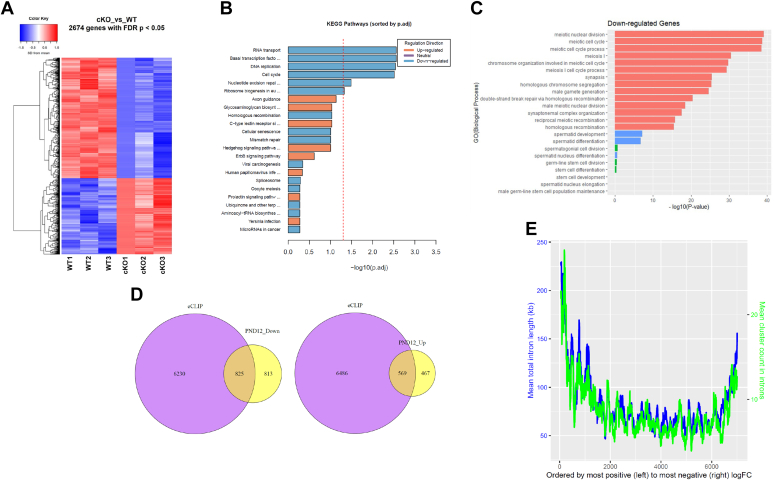
Figure 8.
RNA-Seq and bioinformatics analysis. RNA-Seq to identify differentially expressed genes (DEGs) in Tardbp cKO testis (n = 3) and bioinformatics analysis to identify overlap with previously identified TDP-43 binding RNAs. A, heat map showing 2674 DEGs with 1036 upregulated and 1638 downregulated genes (FDR < 0.05). B, KEGG pathway analysis shows top-enriched pathways for upregulated and downregulated DEGs. C, top-enriched GO biological processes for upregulated and downregulated genes. D, Venn diagram showing comparison of testis DEGs with genes previously identified to have TDP-43 binding sites (CLIP). Approximately 50% of upregulated and 54% of downregulated DEGs contained TDP-43 binding sites. E, correlation between Tardbp cKO testis RNA-Seq and previously published brain TDP-43 CLIP-Seq data. Genes were ranked based on their degree of expression in Tardbp cKO testis (x-axis). The mean total intron length for the next 100 genes was plotted (y-axis, blue line). Likewise, the mean number of intronic CLIP clusters found in the next 100 genes from the ranked list were plotted (y-axis, green line). cKO, conditional KO; FDR, false discovery rate; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and genomes; TDP-43, transactive response DNA-binding protein of 43 kDa.