Figure 5. Use of LC-MS/MS to identify circulating pemphigus anti-desmoglein (dsg) antibodies (abs).
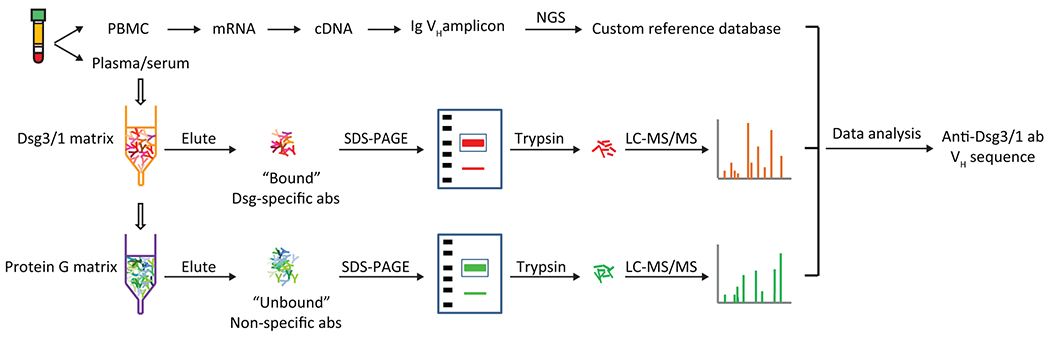
IgG heavy chains from dsg-binding abs and from abs which do not bind to dsg are analyzed by LC-MS/MS. Resultant spectra are compared against a custom database of all variable heavy chain (VH) amino acid (aa) sequences from the same patient to identify ab peptides (for database construction, VH-mRNA transcripts were PCR amplified, sequenced by high-throughput sequencing, and translated into aa to create a VH-specific database of that patient). Peptides that match heavy chain-complementarity region 3 amino acid sequences in the database and that are found only in the bound, but not the unbound pool are informative, allowing definition of the specific clonotype profile in the antigen-specific (dsg-binding) population. NGS, next-generation sequencing; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis. Taken under the CC BY-NC-ND license and under retained rights of the authors from reference Chen et al. (2017).
