Figure 2.
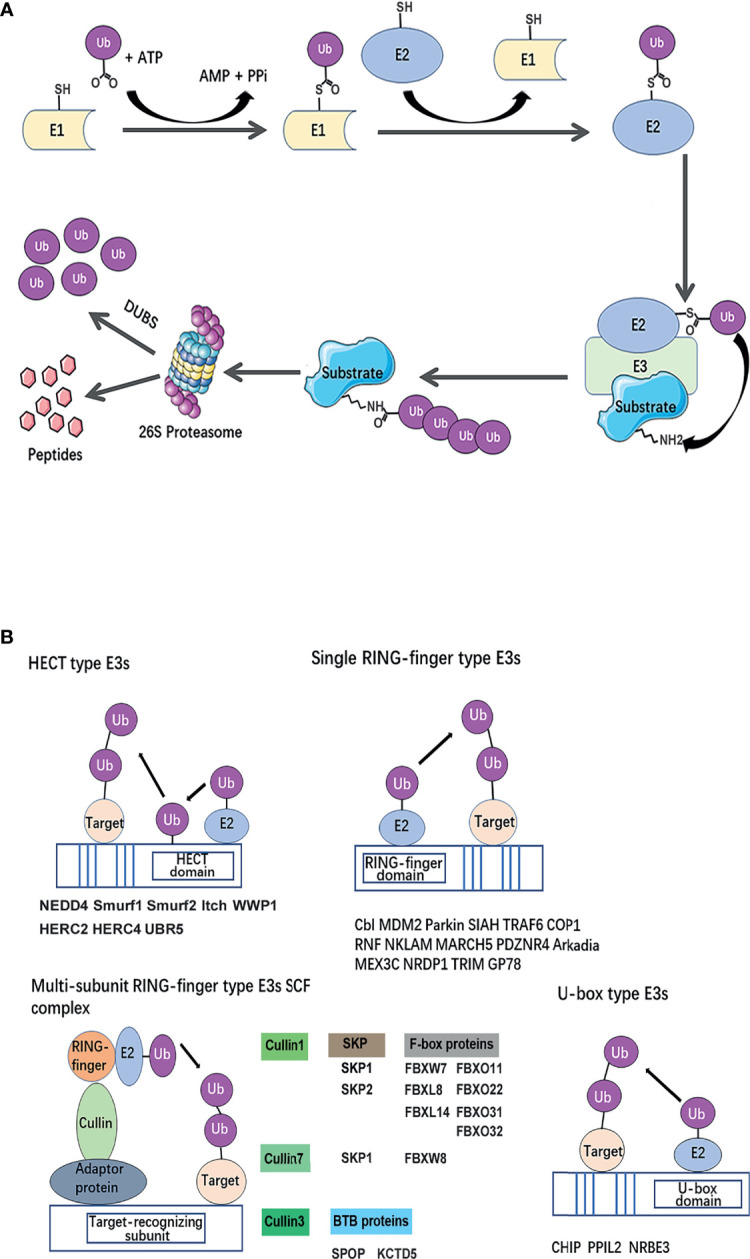
Classical mechanism and structural basis of E3 ubiquitin ligases. (A) The ubiquitin-proteasome system. A ubiquitin-activating enzyme (E1) mediates the activation of the carboxyl-terminal glycine residue of ubiquitin in an ATP-dependent manner. To format the thiolester linkage, the activated ubiquitin is then transferred to E1 followed by the transfer of ubiquitin to a thiolester of a ubiquitin-conjugating enzyme (E2). Ubiquitin protein ligase (E3) confers substrate specificity by recognizing the target proteins and mediating the conjugation of ubiquitin molecules to a lysine residue on the targeted protein via an iso-peptide bond. Polyubiquitinated substrate recognized by 26S proteasome for degradation. (B) Classification of breast cancer metastasis–related ubiquitin E3 ubiquitin ligases based on a structural basis. The canonical E3s are classified into two canonical types: HECT and RING. HECT E3s contains HECT domains which consist of a E2-interacting N-lobe and a catalytic Cys residue containing C-lobe involved in ubiquitin transfer. RING-type E3s mediate the direct transfer of ubiquitin from E2 to substrate, including single RING-finger- type E3s, RING-like (Ubox)-type E3s, and multi-subunit RING-finger-type E3s. The Cullin-RING ubiquitin ligase (CRL) family is composed of a multi-unit. Each type of E3s or members of E3 complexes has been summarized and listed below the schematic diagram of the relevant category.
