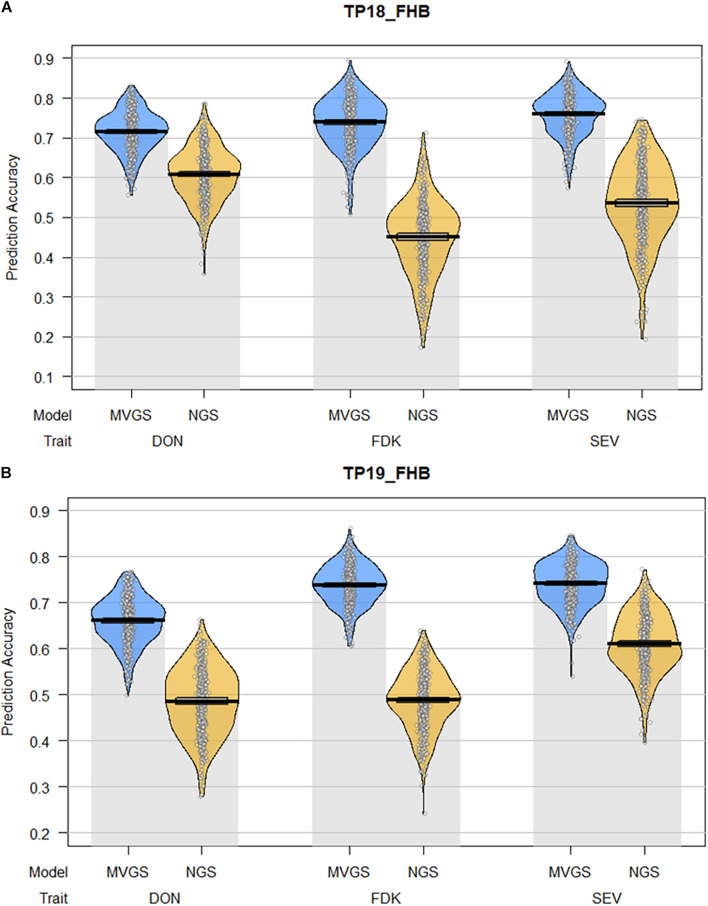
FIGURE 2.
Pirate plots comparing the mean prediction accuracies between multi-trait genomic selection (MTGS) models with naïve genomic selection (NGS) models for three Fusarium head blight resistance traits (FHB), deoxynivalenol (DON) concentration, Fusarium damaged kernels (FDK), and severity (SEV) in soft red winter wheat across two training populations (TPs): (A) TP18_FHB, TP used to predict three FHB resistance traits for the 2018 advanced F4:7 generation (ADV18); (B) TP19_FHB, TP used to predict three FHB resistance traits for the 2019 advanced F4:7 generation (ADV19), consisting of all genotypes from TP18_FHB and ADV18. The x-axis represents the combination of FHB resistance traits and GS model used to predict each trait. The y-axis represents the mean prediction accuracy across 100 iterations of fivefold cross-validation in the form of a Pearson correlation coefficient (r) between the predicted genome-estimated breeding value (GEBV) and the actual phenotypic value for the validation populations. Individual points represent the Pearson correlation from each fold of each iteration of cross-validation for a total of 500 data points. The lines within each plot represent the mean and 95% confidence intervals for prediction accuracy. The curves represent the smoothed densities of the data.