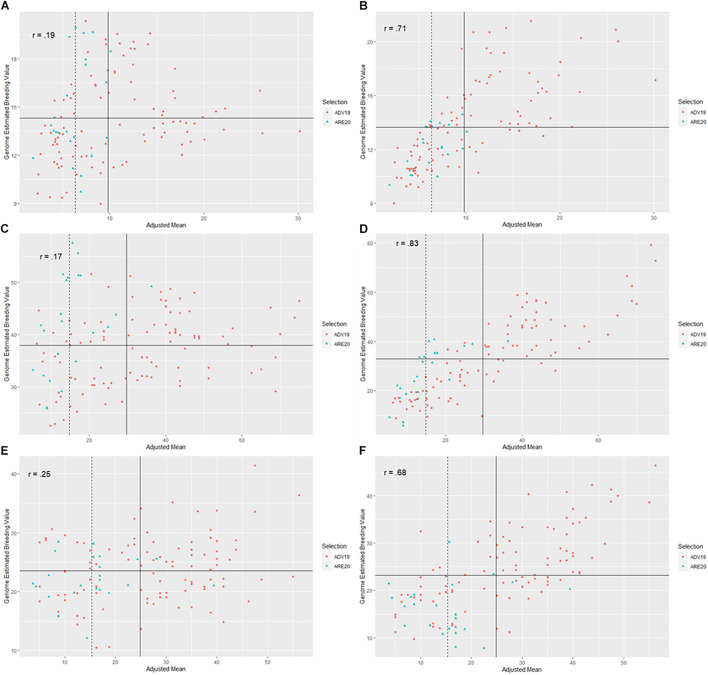
FIGURE 4.
Scatter plots between genome-estimated breeding values (GEBVs) for three Fusarium head blight (FHB) resistance traits in soft red winter wheat from two different genomic selection models (GS), including naïve models without covariates (NGS) and multi-trait GS models with covariates (MTGS), and adjusted means for deoxynivalenol (DON) concentration, Fusarium damaged kernels (FDK), and severity (SEV) across two generations, F4:7 advanced from 2018 to 2019 (ADV19) and F4:8 elite from 2019 to 2020 (ARE20): (A) predictions for DON in ADV19 using a NGS model, (B) predictions for DON using a MTGS model, (C) predictions for FDK from ADV19 using a NGS model, (D) predictions for FDK using a MTGS model, (E) predictions for SEV in ADV18 using a NGS model, (F) predictions for SEV using a MTGS model. The x-axis represents adjusted mean for DON, FDK, or SEV across the ADV and ARE generations. The y-axis represents the GEBVs calculated for DON, FDK, or SEV from the NGS or MTGS models. Different colored data points represent genotypes that were advanced to the next generation. The solid vertical line represents the mean of the adjusted means for the respective FHB resistance trait from the ADV generation, while the vertical dashed line represents the mean of the adjusted means for the respective FHB resistance trait from the ARE generations. The solid horizontal line represents the mean of GEBVs for the respective FHB resistance trait calculated from the NGS or MTGS models. The r label represents the Pearson correlation between GEBVs and adjusted means.