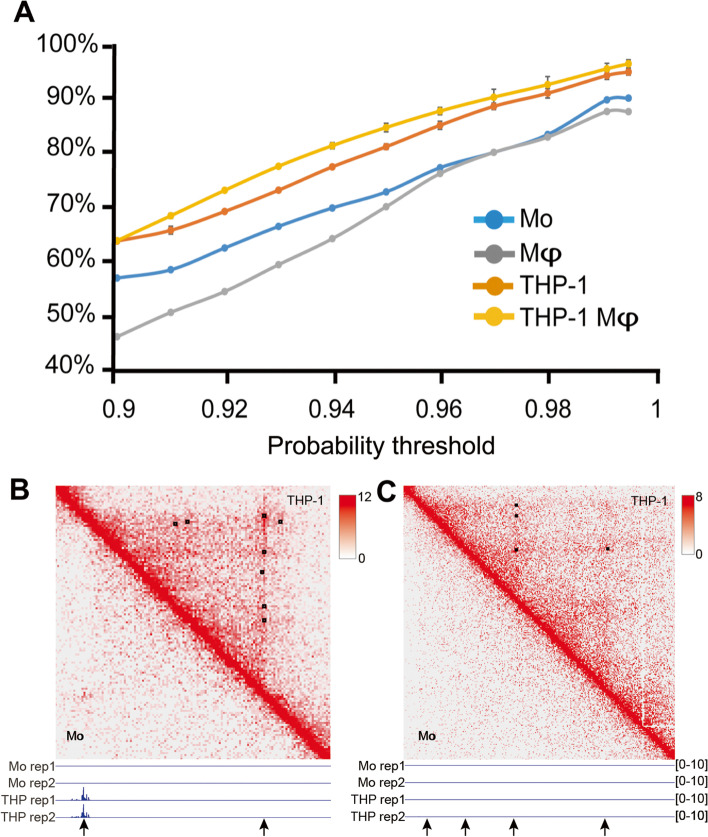
Fig. 2.
Differences in the structure at the loop level between primary and cell lines. a Relationship between the identical loop percentage and the probability threshold cutoff used to filter the loops, comparing all data to the full dataset of the corresponding THP-1 cells. b Hi-C heat map at Chr1:214.7-215.9 M (10 kb resolution) of the monocytic primary and THP-1 cells. The squared regions are the loops identified by Peakachu. The panel at the bottom shows the gene (CENPF) expression in this region, together with the loop anchor locations (arrowed) (c) Hi-C heat map at Chr2:33.8-36.2 M (10 kb resolution) of the monocytic primary and THP-1 cells. Annotations are the same as in (b)