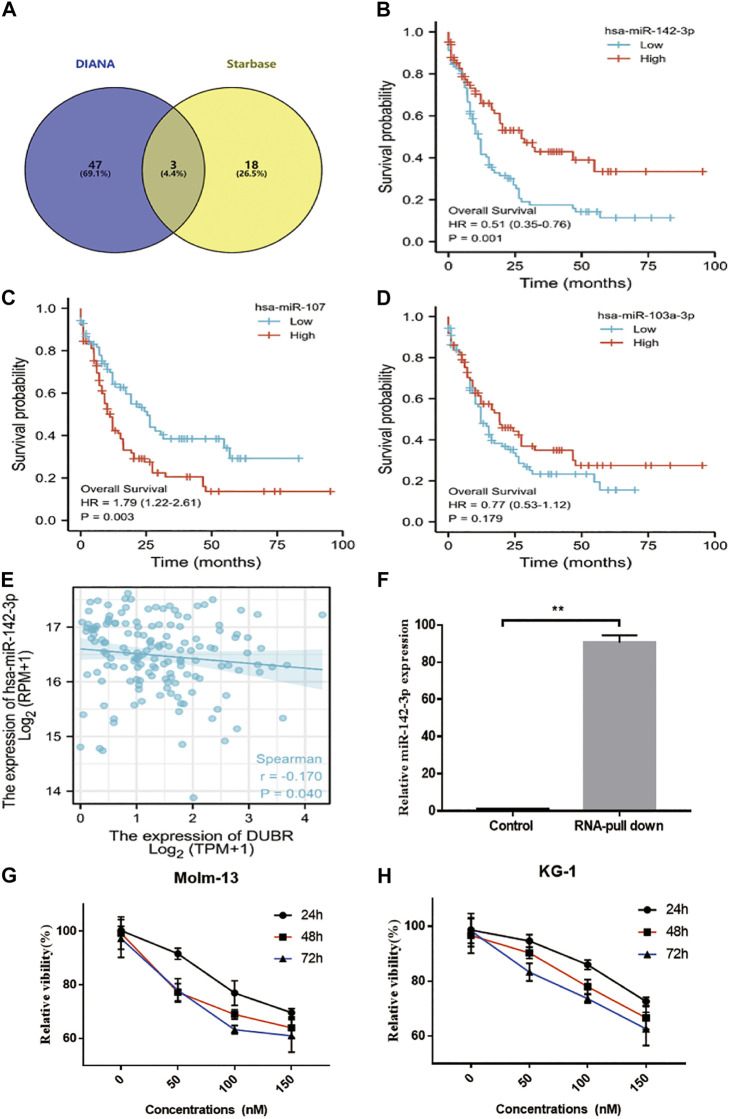
FIGURE 5.
DUBR sponged miR-142–3P and negatively modulated miR-142–3P expression. (A) Starbase2.0 and DIANA were used to predict the DUBR candidate sponge miRNA (miR-142-3P, miR-107, and miR-103-3P were involved in them). The OS plot of miR-142-3P. (B), miR-107 (C) and miR-103-3P (D) in AML. (E) Spearman correlation analysis of relative expression between miR-142-3P and DUBR within AML bone marrow, p < 0.05. (F) RNA pull-down assay and qRT-PCR analysis for the detecting of endogenous miR-142-3P related to the DUBR transcript. SCR or miR-142-3P was transfected into Molm-13 (G) and KG-1 (H) cells. Then, cell viability was measured through CCK8 assay.