FIGURE 1.
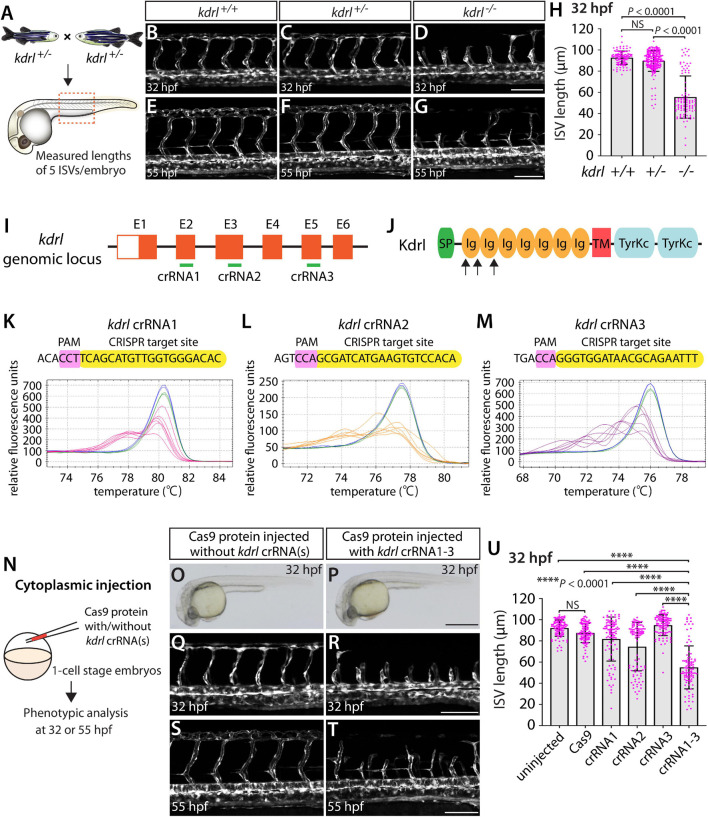
Embryos injected with the triple dgRNPs targeting kdrl exhibited aISV defects similar to those observed in kdrl–/– fish. (A) Experimental setup for the phenotypic analysis presented in panels (B–H). Adult kdrl+ /– fish were incrossed, and progeny generated from the crosses were imaged to quantify the lengths of their arterial intersegmental vessels (aISVs) at 32 hpf. (B–G) Lateral views of kdrl+/+ (B,E), kdrl+ /– (C,F), and kdrl–/– (D,G) trunk vasculature visualized by Tg(kdrl:EGFP) expression at 32 (B–D) and 55 (E–G) hpf. (H) Quantification of aISV lengths in kdrl+/+, kdrl+ /–, and kdrl–/– embryos at 32 hpf (n = 23 for kdrl+/+, n = 45 for kdrl+ /–, and n = 21 for kdrl–/– fish). Lengths of 5 aISVs per embryo were measured. NS: not significant. (I) Three synthetic CRISPR RNAs (crRNAs) were designed to target sequences within exon 2 (E2), E3, and E5 on the kdrl genomic locus. (J) Predicted domain structure of zebrafish Kdrl. Kdrl consists of a signal peptide (SP), seven immunoglobulin-like domains (Ig), a transmembrane domain (TM), and two tyrosine kinase domains (TyrKc). Arrows indicate the approximate positions of the protein sequences corresponding to the target sequences of the three designed crRNAs. (K–M) High-resolution melt analysis (HRMA) used to validate the efficacy of the three designed crRNAs. The melting curves of 6 independent embryos injected with the dgRNP complex containing kdrl crRNA1 (K, pink), crRNA2 (L, orange), or crRNA3 (M, purple) are presented. The melting curves of 2 independent, uninjected (K–M, blue) and Cas9 protein-injected (K–M, green) sibling embryos are also presented for comparison for each crRNA. Injection of each dgRNP cocktail disrupted the corresponding target genomic sequences. (N) Experimental workflow of the microinjection experiments for panels (O–U). Injection cocktails containing Cas9 protein with the individual or the three kdrl crRNA:tracrRNA duplexes were injected into the cytoplasm of one-cell stage Tg(kdrl:EGFP) embryos. Injected Tg(kdrl:EGFP) progeny were analyzed for aISV formation at 32 and 55 hpf. (O,P) Brightfield images of the 32 hpf embryos injected with Cas9 protein with (P) or without (O) the three kdrl crRNA:tracrRNA duplexes (crRNA1-3). (Q–T) Lateral trunk views of the 32 (Q,R) and 55 (S,T) hpf Tg(kdrl:EGFP) fish injected with Cas9 protein with (R,T) or without (Q,S) the three kdrl crRNA1-3. While aISVs in Cas9-injected embryos fused at the dorsal side of the trunk to form the dorsal longitudinal anastomotic vessel by 32 hpf (Q), a vast majority of aISVs in the embryos injected with the triple kdrl dgRNPs failed to reach the dorsal side of the trunk (R). This aISV stalling phenotype was observed at later developmental stages, including at 55 hpf (T). (U) Quantification of aISV lengths of 32 hpf embryos of indicated treatment (n = 20 fish for each treatment group; 5 aISV lengths per embryo measured). Average aISV lengths of the triple dgRNPs-injected embryos closely resembled those of kdrl–/– fish. A one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test was used to calculate P values for panels (H,U). Scale bars: 50 μm in (D,G,R,T); 500 μm in (P).