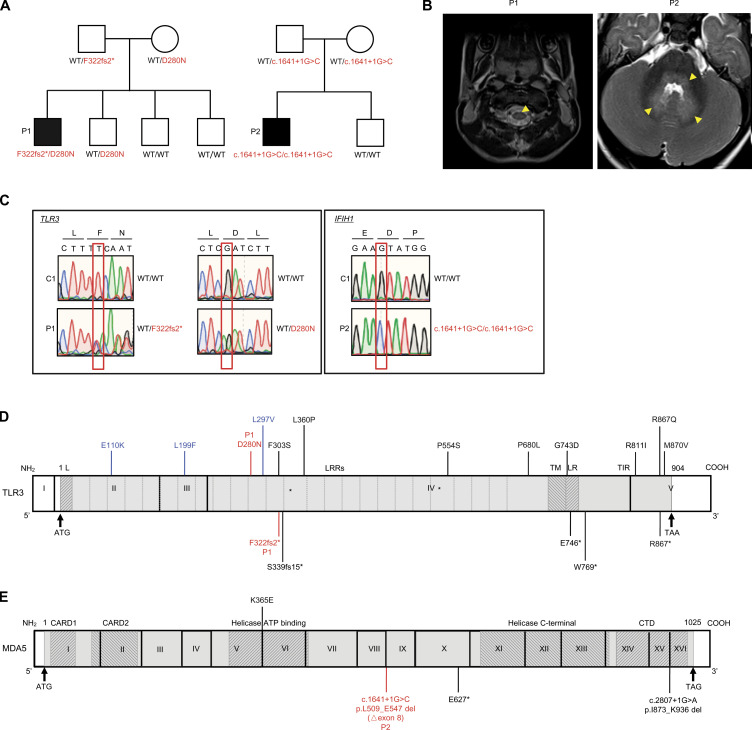
Figure 1.
Biallelic TLR3 or IFIH1 mutations in two patients with EVE. (A) Family pedigrees, showing segregation of the TLR3 and IFIH1 mutations. The black symbols indicate patients. (B) Brain imaging showing EVE lesions in P1 and P2. In P1, post-contrast T1-FLAIR imaging showed pathological tegmental hypersignal (yellow arrows) associated with cervical myelitis, highly suggestive of rhombencephalitis. In P2, post-contrast T1-FLAIR imaging showed pathological tegmental hypersignal with evidence of diffusion restriction in the superior cerebellar peduncles and the ponto-bulbar junction (yellow arrows), suggestive of rhombencephalitis. (C) Confirmation of the mutations by Sanger sequencing. The single nucleotide substitutions for each patient relative to the sequence for a healthy control are indicated by red squares. (D) Schematic diagram of the structure of the human TLR3 gene and protein, featuring the leader sequence (L), leucine-rich repeats (LRRs), transmembrane domain (TM), linker region (LR), and Toll/IL-1 receptor (TIR) domain. Roman numerals indicate the coding exons. Mutations previously identified in patients with HSE (E110K, L297V, L360P, P554S, G743D, E746*, R811I, R867Q), IAV or SARS-CoV-2 pneumonia (S339fs15*, P554S, P680L, W769*, M870V) or patients with other viral infections (F303S, L199F, R867*) are shown in black (experimentally characterized) or blue (not characterized). The mutations (D280N and F322fs2*) found in P1 are shown in red. (E) Schematic diagram of the structure of the human IFIH1 gene and MDA5 protein, featuring the CARD domain, helicase ATP-binding domain, helicase C-terminal domain, and carboxy-terminal domain. Roman numerals indicate the coding exons. Mutations previously identified in patients with respiratory virus infections are shown in black. The mutation (c.1641+1G>C, p.L509_E547 del [△exon8]) found in P2 is shown in red.