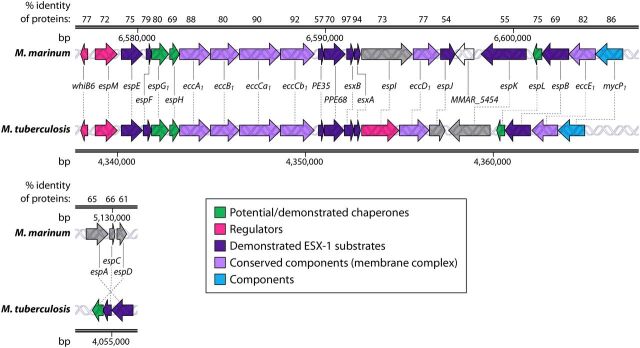
FIG 2.
Syntenic esx-1 loci from M. marinum and M. tuberculosis. The genes and their locations in each genome as well as the percent amino acid identities of the resulting proteins from the conserved esx-1 and espACD loci in M. marinum and M. tuberculosis are shown. MMAR_5454 is a gene found only in M. marinum. Gray shading indicates that the gene function has not been defined. Differences in colors between organisms, for example, for PPE68, indicate that PPE68 is a substrate in M. marinum but is required for secretion and not yet classified as a substrate in M. tuberculosis. The RD1 region is shown (7), as follows. ESX-1 gene products from M. tuberculosis (GenBank accession number NC_000962.3) were searched against those from Mycobacterium marinum (NCBI taxonomy accession number txid1781) using NCBI BLASTP (169). The percent identities of the top orthologous proteins in M. marinum are reported at the top.