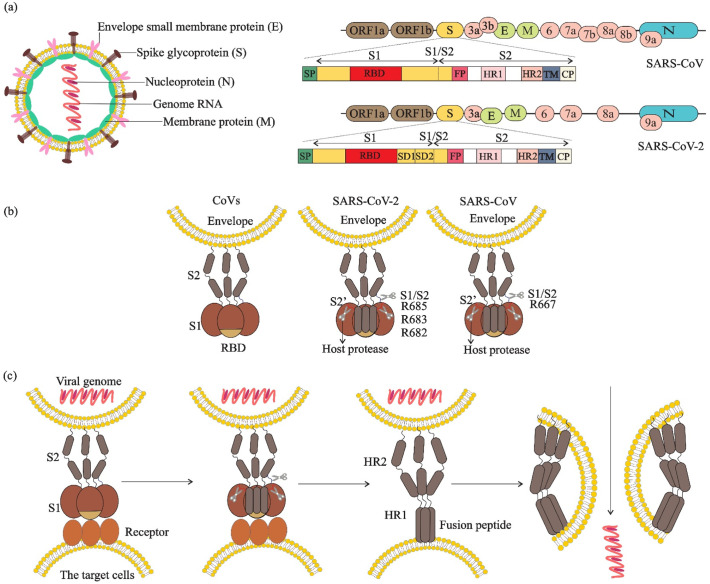
Fig. 1.
Virus particles, complete genome sequences, spike structure, cleavage sites and fusion reaction of SARS-CoV and SARS-CoV-2. a) Virions contain a +ssRNA genome of 26–32 kb in size. The genome ORF1a/b encodes polyproteins, which form the viral replicase transcriptase complex. The other ORFs on the genome encode four main structural proteins: S, E, N and M proteins, as well as several accessory proteins. b) The spike protein structure is composed of an extracellular (EC) domain, a transmembrane anchor domain and a short intracellular tail. EC domain contains two functional subunits, a receptor-binding subunit (S1) and a membrane-fusion subunit (S2), S1 contains two independent domains, an N-terminal domain (S1-NTD) and receptor binding domain (RBD), the S1/S2 cleavage site is shown in its uncleaved, native state and resides in an unstructured region between S1 and S2, the S2’ cleavage site is exposed only after receptor binding. c) Fusion reaction; S1 attaches to a receptor on the target cells, induces a conformational change in the S, exposing cleavage sites between S1 and S2. In SARS-CoV-2, the trimeric S protein then cleaves into S1 and S2 subunits by cellular proteases (scissors), the fusion peptide (FP) latches onto the target membrane, anchoring the virus and cell together. The heptad repeat 2 (HR2) then folds to interact with the heptad repeat 1 (HR1), bringing the membranes together. The successful refolding of enough adjacent S2s leads to fusion of viral and cell membranes and release of the viral genome into the target cell cytoplasm.