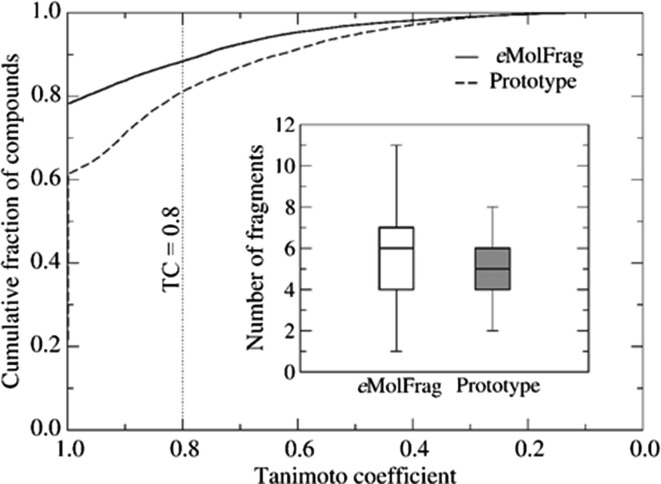
Figure 3.
Bioactive compounds from the Database of Useful (Docking) Decoys Enhanced (DUD-E) database fragmented with eMolFrag. eMolfrag was able to generate an average of six fragments per molecule. eSynth uses beam search techniques to create new drug molecules by combining the building blocks generated by eMolFrag in a chemically comprehensive way. By using fragments generated by eMolFrag, eSynth reconstructed 78.3% of active compounds with a Tanimoto coefficient (TC) of 1.0 and 88.4% with a TC ≥ 0.8. Adapted from Liu et al.26