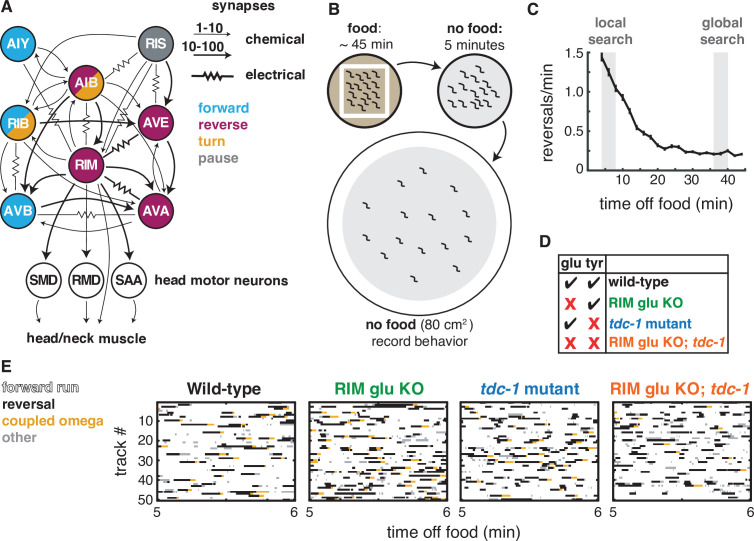
Figure 1. Two RIM neurotransmitters affect spontaneous locomotion.
(A) RIM synapses with interneurons, motor neurons, and muscle implicated in spontaneous foraging behavior (Cook et al., 2019; White et al., 1986). Colors of neurons indicate associated locomotor states based on neural manipulations and functional calcium imaging (Alkema et al., 2005; Gray et al., 2005; Kato et al., 2015; Li et al., 2014; Pokala et al., 2014; Steuer Costa et al., 2019; Wang et al., 2020; Zheng et al., 1999). (B) Off-food foraging assay. (C) Mean reversals per minute of wild-type animals in foraging assays. Vertical lines indicate standard error of the mean. Gray shaded boxes indicate local search (4–8 min off food) and global search (36–40 min off food) intervals analyzed in subsequent figures. n = 324. (D) RIM neurotransmitter mutants. RIM glu KO: RIM-specific knockout of the vesicular glutamate transporter EAT-4 (Figure 1—figure supplement 1). tdc-1, tyrosine decarboxylase mutant, which lacks tyramine in RIM and octopamine in RIC. (E) Ethograms of 50 randomly chosen tracks per genotype during minute 5–6 of local search. Color code: white, forward runs; black, reversals; yellow, omega turns coupled to a reversal; gray, pauses, shallow turns, and omega turns that were not preceded by a reversal.