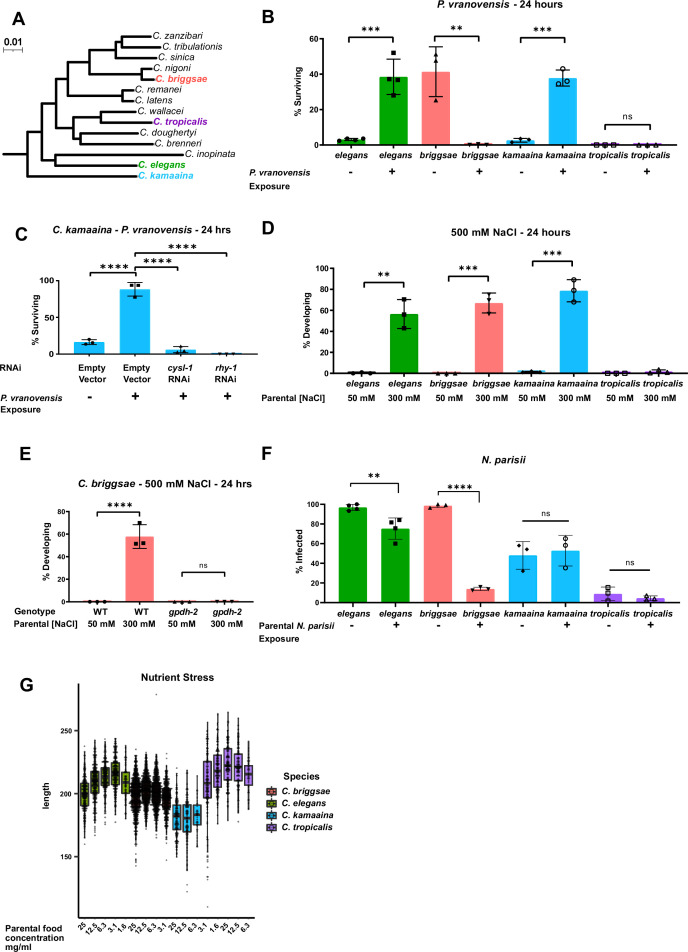
Figure 1. Intergenerational adaptations to multiple stresses are evolutionarily conserved in multiple species of Caenorhabditis.
(A) Phylogenetic tree of the Elegans group of Caenorhabditis species adapted from Stevens et al., 2020. Scale represents substitutions per site. (B) Percent of wild-type C. elegans (N2), C. kamaaina (QG122), C. briggsae (AF16), and C. tropicalis (JU1373) animals surviving after 24 hr on plates seeded with P. vranovensis BIGb0446. Data presented as mean values ± s.d. n = 3–4 experiments of >100 animals. (C) Percent of C. kamaaina wild-type (QG122) animals surviving after 24 hr of exposure to P. vranovensis. Data presented as mean values ± s.d. n = 3 experiments of >100 animals. (D) Percent of wild-type animals mobile and developing at 500 mM NaCl after 24 hr. Data presented as mean values ± s.d. n = 3 experiments of >100 animals. (E) Percent of wild-type and Cbr-gpdh-2(syb2973) mutant C. briggsae (AF16) mobile and developing after 24 hr at 500 mM NaCl. Data presented as mean values ± s.d. n = 3 experiments of >100 animals. (F) Percent of animals exhibiting detectable infection by N. parisii as determined by DY96 staining after 72 hr for C. elegans and C. briggsae, or 96 hr for C. kamaaina and C. tropicalis. Data presented as mean values ± s.e.m. n = 3–4 experiments of 83–202 animals. (G) Boxplots for length of L1 progeny from P0 parents that were subject to the HB101 dose series. Larvae were measured using Wormsizer. Boxplots show median length with four quartiles. n = 3–8 experiments of 50–200 animals. **p < 0.01, ***p < 0.0001, ****p < 0.0001.