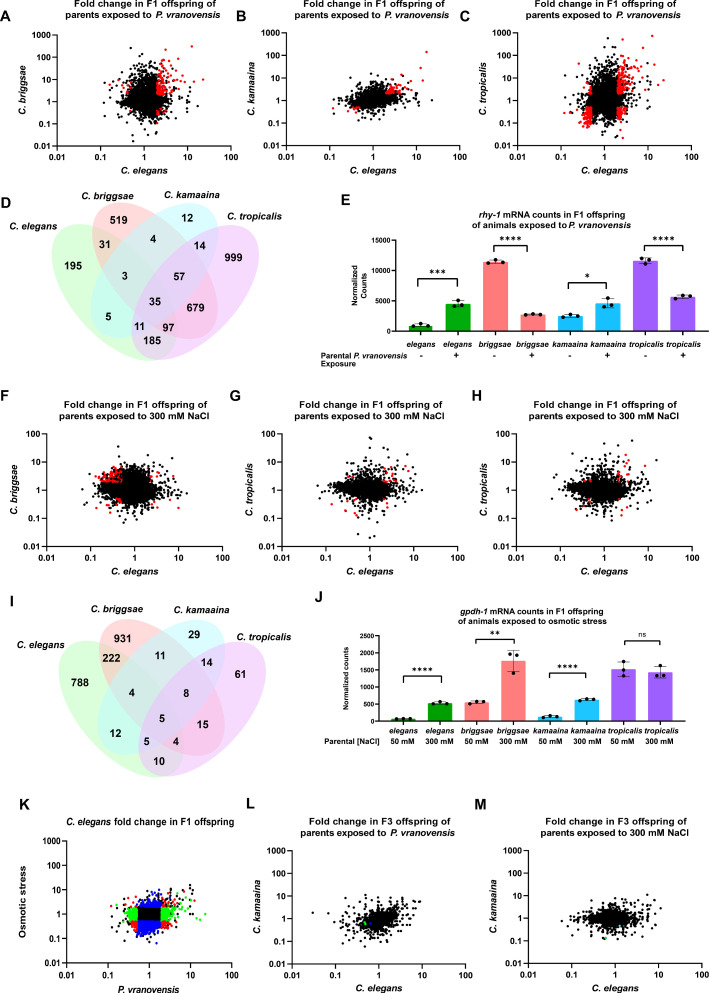
Figure 2. Parental exposure to P. vranovensis and osmotic stress have overlapping effects on offspring gene expression across multiple species.
(A) Average fold change of 7587 single-copy ortholog genes in F1 progeny of C. elegans and C. briggsae parents fed P. vranovensis BIGb0446 when compared to parents fed E. coli HB101. Average fold change from three replicates. Red dots represent genes that exhibit >twofold (padj <0.01) changes in expression in both species. (B) Average fold change of 7587 single-copy ortholog genes in F1 progeny of C. elegans and C. kamaaina parents fed P. vranovensis BIGb0446 when compared to parents fed E. coli HB101. Average fold change from three replicates. Red dots represent genes that exhibit >twofold (padj <0.01) changes in expression in both species. (C) Average fold change of 7587 single-copy ortholog genes in F1 progeny of C. elegans and C. tropicalis parents fed P. vranovensis BIGb0446 when compared to parents fed E. coli HB101. Average fold change from three replicates. Red dots represent genes that exhibit > twofold (padj <0.01) changes in expression in both species (D) Venn diagram of the number of genes that exhibit overlapping >2 fold (padj <0.01) changes in expression in F1 progeny of animals exposed to P. vranovensis BIGb0446 in each species. (E) Normalized counts of reads matching orthologs of rhy-1 in the F1 offspring of parents fed either E. coli HB101 or P. vranovensis BIGb0446. Data from Supplementary file 2. n = 3 replicates. (F) Average fold change of 7587 single-copy ortholog genes in F1 progeny of C. elegans and C. briggsae parents grown at 300 mM NaCl when compared to parents grown at 50 mM NaCl. Average fold change from three replicates. Red dots represent genes that exhibit >twofold (padj <0.01) changes in expression in both species. (G) Average fold change of 7587 single-copy ortholog genes in F1 progeny of C. elegans and C. kamaaina parents grown at 300 mM NaCl when compared to parents grown at 50 mM NaCl. Average fold change from three replicates. Red dots represent genes that exhibit >twofold (padj <0.01) changes in expression in both species in both species. (H) Average fold change of 7587 single-copy ortholog genes in F1 progeny of C. elegans and C. tropicalis parents grown at 300 mM NaCl when compared to parents grown at 50 mM NaCl. Average fold change from three replicates. Red dots represent genes exhibit >twofold (padj <0.01) changes in expression in both species. (I) Venn diagram of the number of genes that exhibit overlapping >twofold (padj <0.01) changes in expression in F1 progeny of animals grown at 300 mM NaCl in each species. (J) Normalized counts of reads matching orthologs of gpdh-1 in the F1 progeny of parents grown at either 300 mM NaCl or 50 mM NaCl. Data from Supplementary file 3. n = 3 replicates. (K) Average fold change for 7587 ortholog genes in F1 progeny of C. elegans parents fed P. vranovensis or exposed to 300 mM NaCl when compared to naive parents. Average fold change from three replicates. Red dots – genes that change in expression in response to both stresses. Blue dots – genes that change in expression in response to only osmotic stress. Green dots – genes that change in expression in response to only P. vranovensis. (L) Average fold change of 7512 single-copy ortholog genes in F3 progeny of C. elegans and C. kamaaina fed P. vranovensis BIGb0446 when compared to those fed E. coli HB101. Average fold change from three replicates. Blue dots represent genes that exhibited >twofold (padj <0.01) changes in expression in C. elegans. Green dots represent genes that exhibited >twofold (padj <0.01) changes in expression in C. kamaaina. (M) Average fold change of 7512 single-copy ortholog genes in F1 progeny of C. elegans and C. kamaaina parents grown at 300 mM NaCl when compared to parents grown at 50 mM NaCl. Average fold change from three replicates. Green dots represent genes that exhibited >twofold (padj <0.01) changes in expression in C. kamaaina. *p < 0.05, **p < 0.01, ***p < 0.0001, ****p < 0.0001.