Figure 3. Protein arginine methyltransferases regulate the p53 and DNA damage response pathways.
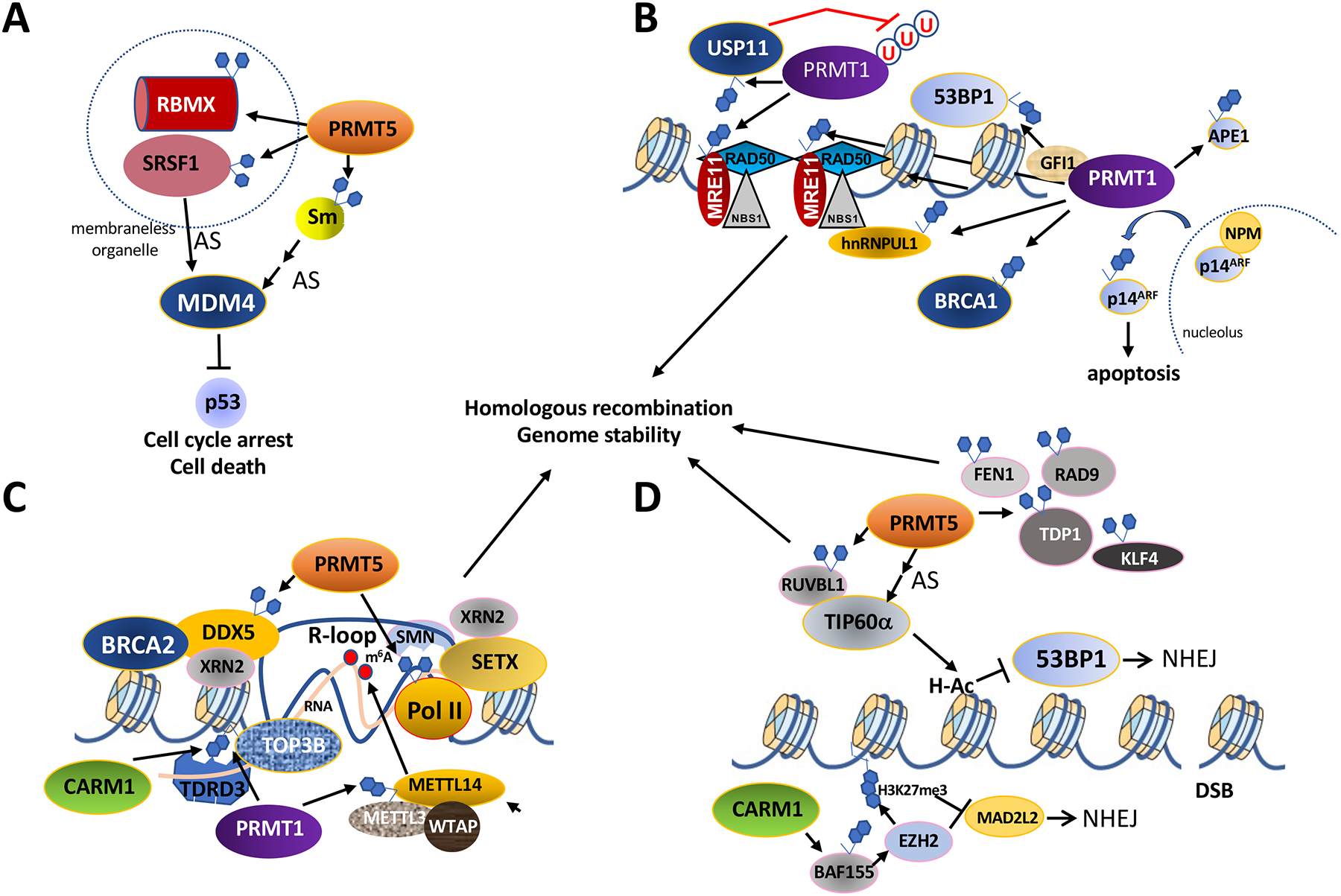
A. PRMT5 methylates Sm proteins, SRSF1 (Serine and arginine rich splicing factor 1), and RBMX (RNA binding motif protein X-linked), to regulate MDM4 alternative splicing (AS) and p53 protein levels. SRSF1 and RBMX can coexist in a membraneless nuclear organelle and this is regulated by the methylation of RBMX by PRMT5.
B. The MRE11-RAD50-NBS1 sensor complex at DSBs (double stranded breaks) activates the ATM (Ataxia telangiectasia mutated) kinase to phosphorylate the histone variant H2AX (γH2AX) and many other proteins to trigger DSB repair. PRMT1 methylation of MRE11 regulates its resection activities. The methylation of RNA binding protein hnRNPUL1 regulates interaction with NBS1. GFI1 is a transcription factor that enhances the PRMT1-mediated methylation of MRE11 in T cells and USP11 regulates the activity of PRMT1 by deubiquination. Circled ‘U’ denote ubiquitin. The methylation of the DSB repair proteins 53BP1, BRCA1, APE1 (apurinic/apyrimidinic endonuclease 1) is mediated by PRMT1, but the role of these methylation events remain unknown. Methylation of p14ARF (alternative reading frame tumor suppressor product of the CDKN2A locus) allows its nucleolar export from NPM (nucleophosmin) to promoter apoptosis.
C. DDX5 (DEAD-box helicase 5) is methylated by PRMT5 and this regulates association with XRN2 (5’−3’ exonuclease 2) for R-loop (RNA/DNA hybrid) resolution. DDX5 recruitment to DNA breaks is regulated by BRCA2 (Breast cancer gene 2). PRMT5 also methylates the RNA polymerase II C-terminal domain which lead to the recruitment of the helicase SETX (Senataxin) and XRN2 by SMN to resolve R-loops. CARM1 and PRMT1 methylates histones which attract the methyl-reader protein TDRD3 via its Tudor domain to recruit topoisomerase TOP3B (DNA topoisomerase III beta) to resolve R-loops. The resolution of R-loops prevents endogenous DNA damage and maintains genome stability. The RNA of the RNA/DNA hybrid of R-loops harbors m6A (N6-methyladenosine) and this event is regulated by PRMT1 methylation of the m6A methyltransferase (WTAP, METTL3, METTL14) component METTL14.
D. PRMT5 methylates RUVBL1 (RuvB like AAA ATPase) activating TIP60α which acetylates H4K16 to block 53BP1 recruitment favoring HR (homologous recombination) over NHEJ (non-homologous end-joining). The alternative splicing (AS) of TIP60α is regulated by PRMT5 to promote HR. CARM1 methylation of the SWI/SNF complex subunit BAF155 at R1064 increases EZH2 (Enhancer of zeste 2 polycomb repressive complex 2 subunit) H3K27me3 methylation repressing NHEJ Shieldin component MAD2L2. The upregulation of NHEJ in HR-proficient high-grade serous ovarian cancers by EZH2 inhibition creates sensitivity to poly (ADP-ribose) polymerase inhibitors. PRMT5 also methylates FEN1, RAD9, TDP1, and KLF4 to maintain genomic stability.
