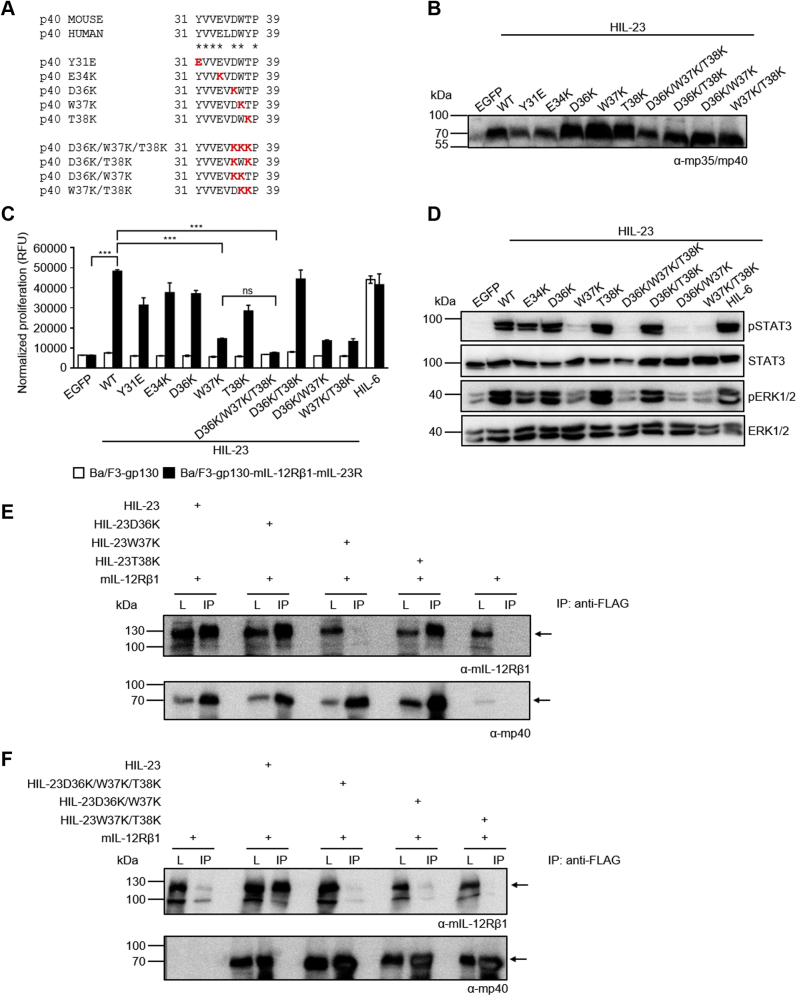
Figure 2.
W37 of murine p40 is important for binding of HIL-23 to murine IL-12Rβ1.A, alignment of murine and human p40 N-terminal amino acids Y31 to P39. Single, double, and triple substitutions within mp40 are highlighted in red. B, Western blot analysis of secreted murine HIL-23 variants from transfected CHO-K1 cells. C, cellular proliferation of Ba/F3-gp130-mIL-12Rβ1-mIL-23R cells. The cells were cultured for 3 days in the presence of 10 ng/ml HIL-6 or with the indicated cytokines (10% conditioned cell culture supernatant of transfected CHO-K1 cells). Parental Ba/F3-gp130 cells were used as controls. The results of one representative experiment of three are shown. Error bars represent S.D. for technical replicates. Statistical analysis used a one-way ANOVA, followed by Bonferroni correction (n = 3), ∗∗∗p ≤ 0.001, ns not significant. D, analysis of STAT3 and ERK1/2 activation. Ba/F3-gp130-mIL-12Rβ1-mIL-23R cells were washed, starved, and stimulated with the indicated cytokines (10% conditioned cell culture supernatant of transfected CHO-K1 cells) for 30 min. Cellular lysates were prepared, and equal amounts of total protein (50 μg/lane) were loaded on SDS-PAA gels, followed by immunoblotting using specific antibodies for phospho-STAT3, STAT3, phospho-ERK1/2, and ERK1/2. Western blotting data show results of one representative experiment of three. E, co-IP of FLAG-tagged murine HIL-23 variants (wild-type, D36K, W37K and T38K) and full-length mIL-12Rβ1. The position of mIL-12Rβ1 and HIL-23 variants is indicated by arrows. One of two independent experiments is shown. F, co-IP of FLAG-tagged murine HIL-23 variants (wild-type, D36K/W37K/T38K, D36K/W37K, W37K/T38K) and full-length mIL-12Rβ1. The position of mIL-12Rβ1 and HIL-23 variants is indicated by arrows. One of two independent experiments is shown.