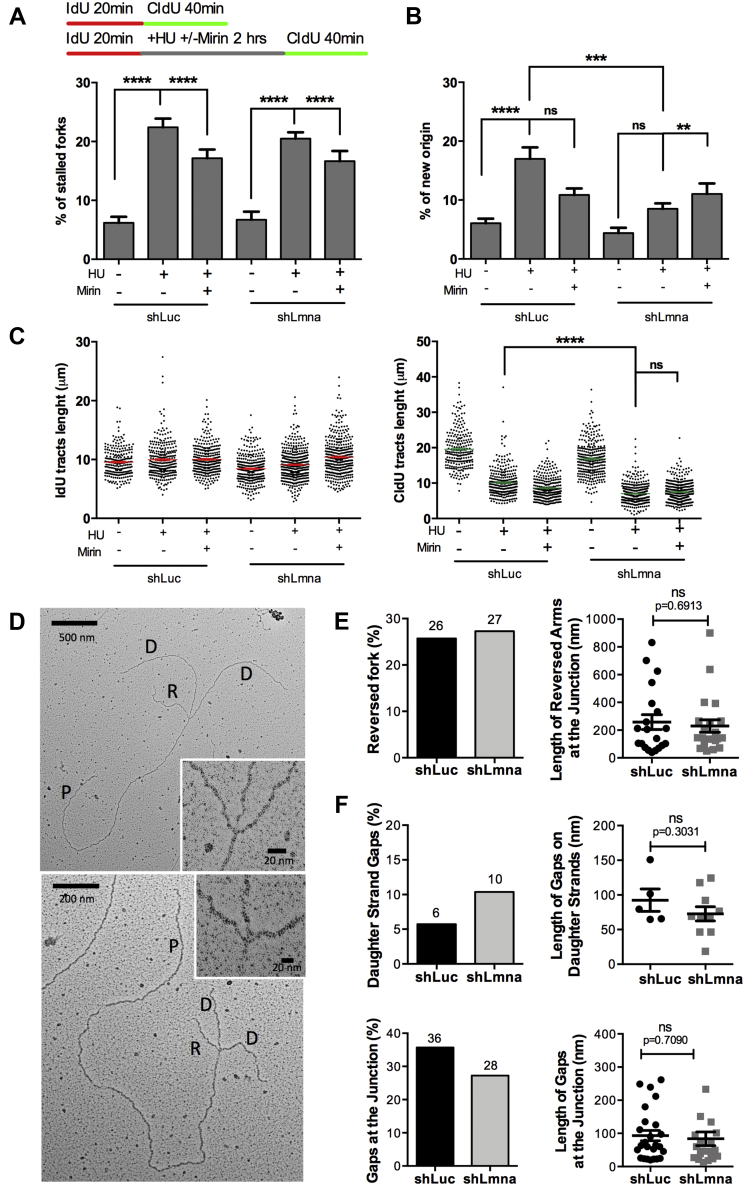
Figure 4.
RFI in lamin A/C-depleted cells is independent of fork reversal.A, HEK-293T cells depleted of lamin A/C (shLmna) or control cells (shLuc) were used for DNA fiber assays with a labeling scheme that monitors fork restart. Cells are labeled with IdU (red) for 20 min, then arrested with HU (4 mM for 2 h) in the presence or the absence of Mirin, and released into complete media with CldU (green) for 40 min to label DNA after fork restart. A control sample was included without any drugs (IdU 20 min + CldU 40 min). The percentage of stalled forks (only red label) is quantified in the graph. B, the same labeling scheme as in (A) and new origins (green only) are quantified. C, same labeling scheme as in (A), and measurement of IdU tracts length (left graph), a measure of replication rate, and CldU tracts length (right graph), a measure of fork restart. Note how lamin A/C depletion hinders fork restart. Graph shows each individual value of all measurements from all three biological repeats. The bar indicates the average of all individual measurements ± SEM. D, EM performed in lamin A/C-proficient and lamin A/C-deficient cells to visualize reversed forks and extent of ssDNA gaps at the stalled fork. The cells were treated with HU (4 mM HU for 2 h) to induce RS and RFI in lamin A/C-depleted cells and with Mirin (50 μM for 2 h) to prevent degradation of reversed forks. RF structures are identified as two arms of equal length and reversed forks as four-way junction structures. Two examples of reversed forks in lamin A/C-depleted HEK-293T cells are shown. E, frequency of fork reversal in lamin A/C-proficient and lamin A/C-deficient cells treated with HU and Mirin (left graph) and length (nanometer) of the reversed arms at the junction. Note that there are no differences between control (sLuc) and lamin A/C-deficient (shLmna) cells. F, frequency of ssDNA gaps in daughter strands and at the junction (left graphs) and length of the gaps (right graphs) in lamin A/C-depleted and control cells. CldU, chlorodeoxyuridine; HEK-293T, human embryonic kidney 293T cells; HU, hydroxyurea; IdU, iododeoxyuridine; RFI, replication fork instability; RS, replication stress.