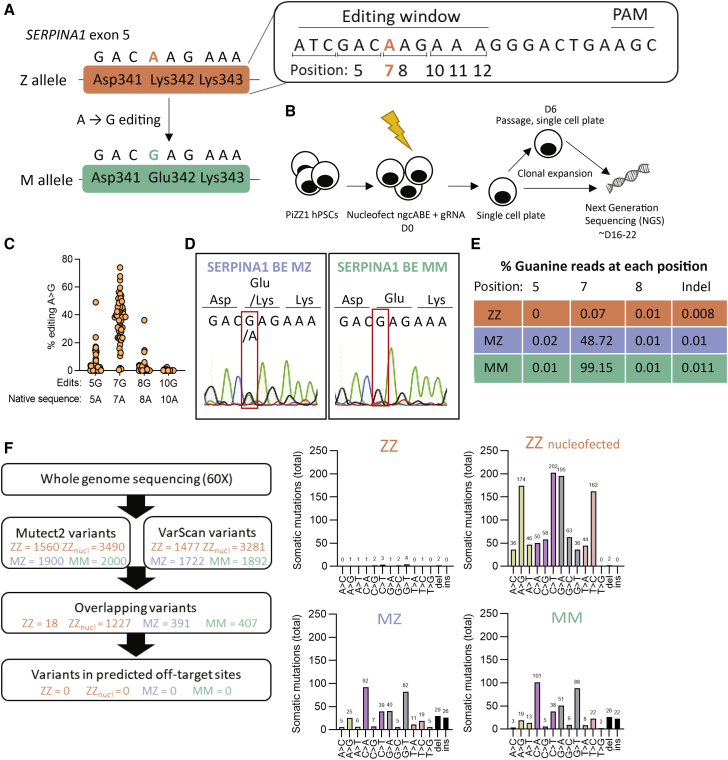
Figure 1.
Base editing corrects the Z mutation in AATD iPSCs
(A) Z allele (Lys342) and desired A > G edit to M allele. (B) Schematic representation of workflow. iPSCs (PiZZ1 line) were nucleofected with adenine base editor (ngcABEvar5) and guide RNA (gRNA), then immediately plated as single cells. Cells were allowed to clonally expand, and some colonies were passaged again to single cells at D6 post-nucleofection. Clones were collected and analyzed using next-generation sequencing (NGS). (C) Editing efficiency of base editor to correct Z mutation. Fifty-six clones, each clone represented by a circle, were screened by NGS for on-target editing at the Z mutation (A > G) and for bystander edits within the editing window. (D) Following additional subcloning, pure populations of MZ and MM iPSCs were obtained and analyzed using Sanger sequencing. (E) NGS of pure ZZ, MZ, and MM iPSCs showing percentage of reads at each position. (F) Whole-genome sequencing was performed with 60× coverage. Passage-matched samples (ZZ, ZZ cells that were nucleofected without editors, MZ, and MM) were compared with earlier passage ZZ cells (the same passage used to commence base editing). Variants were detected using Mutect2 and VarScan, then these variants were overlayed with predicted off-target mutation sites (determined with COSMID), with no mutations overlapping with off-target mutation sites.