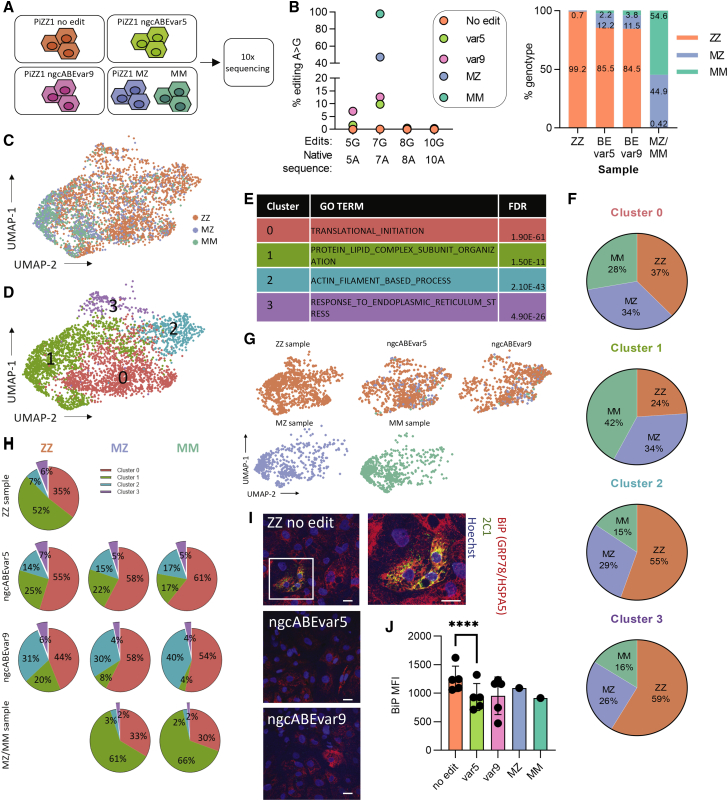
Figure 5.
Base-edited iHeps are protected from ER stress.
(A) Schematic representation of 10x Genomics single-cell RNA sequencing (scRNA-seq) experiment. (B) Editing efficiency was determined using DNA next-generation sequencing (left) or Vartrix Single-Cell Genotyping tool (10x Genomics) (right). (C) UMAP representation of ZZ, MZ, and MM genotyped cells. (D) UMAP visualization of Louvain clusters at a resolution of 0.17. (E) Gene set enrichment analysis of differentially expressed genes was determined for each Louvain cluster with the top GO biological process (BP) term shown. (F) Proportion of each Louvain cluster by ZZ, MZ, and MM genotypes, normalized by total number of cells. (G) UMAP projections for each sample showing ZZ, MZ, and MM genotyped cells. (H) Proportion of each genotype in Louvain clusters in each sample. (I) Immunofluorescent staining for binding immunoglobulin protein (BiP; also called HSPA5 or GRP78) (red), 2C1 (polymerized AAT) (green), and Hoechst (blue). Representative images from one of three experiments are shown; scale bar, 20 μm. (J) BiP mean fluorescent intensity (MFI) was quantified using flow cytometry; n = 5 independent differentiations; error bars represent SD, ∗∗∗∗ p ≤ 0.0001.