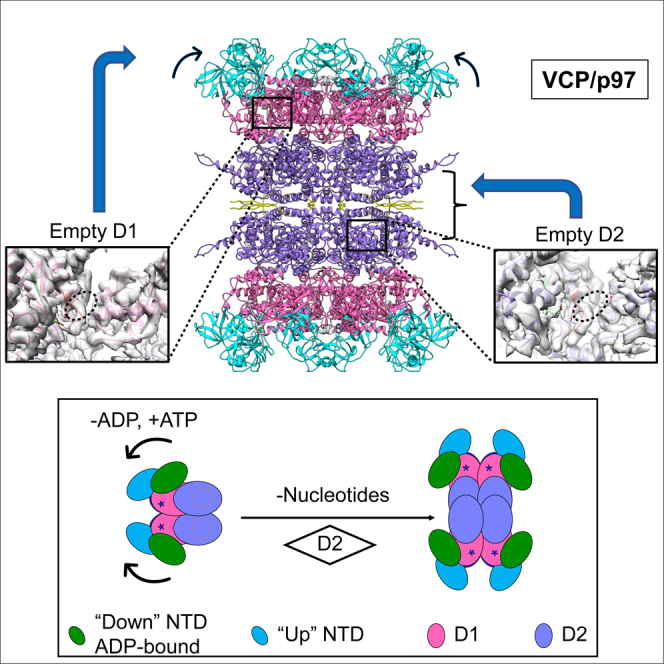
Summary
VCP/p97 is an evolutionarily conserved AAA+ ATPase important for cellular homeostasis. Previous studies suggest that VCP predominantly exists as a homohexamer. Here, we performed structural and biochemical characterization of VCP dodecamer, an understudied state of VCP. The structure revealed an apo nucleotide status that has rarely been captured, a tail-to-tail assembly of two hexamers, and the up-elevated N-terminal domains akin to that seen in the ATP-bound hexamer. Further analyses elucidated a nucleotide status-dependent dodecamerization mechanism, where nucleotide dissociation from the D2 AAA domains induces and promotes VCP dodecamerization. In contrast, nucleotide-free D1 AAA domains are associated with the up-rotation of N-terminal domains, which may prime D1 for ATP binding. These results therefore reveal new nucleotide status-dictated intra- and interhexamer conformational changes and suggest that modulation of D2 domain nucleotide occupancy may serve as a mechanism in controlling VCP oligomeric states.
Subject areas: Biological sciences, Biochemistry, Structural biology
Graphical abstract
Highlights
-
•
A cryo-EM structure of VCP dodecamer purified with mammalian cells is resolved
-
•
The dodecamer structure reveals an apo nucleotide status
-
•
A VCP dodecamerization mechanism has been elucidated
-
•
Loss of nucleotide from D2 promotes VCP dodecamer formation
Biological sciences; Biochemistry; Structural biology
Introduction
Valosin-containing protein (VCP, also known as p97 in mammals and Cdc48 in yeast) is an evolutionarily conserved type II AAA+ (ATPases Associated with diverse cellular Activities) ATPase, which plays an essential role in the maintenance of cellular homeostasis by orchestrating protein turnover through either the ubiquitin-proteasome system (UPS) or the autophagy/lysosomal route (Meyer et al., 2012; van den Boom and Meyer, 2018). By processing a wide spectrum of ubiquitinated substrates, it is implicated in a plethora of cellular processes ranging from degradation of damaged proteins and organelles to chromatin remodeling, DNA damage repair, endosomal trafficking, and homotypic membrane fusion in the biogenesis of organelles (Meyer et al., 2012). Mutations or changes in VCP expression or activity are linked to multiple degenerative disorders (Tang and Xia, 2016; Meyer and Weihl, 2014) and cancer (Anderson et al., 2015; Deshaies, 2014; Magnaghi et al., 2013), making VCP an important therapeutic target.
VCP contains an N-terminal domain (NTD), two tandem AAA+ ATPase domains (D1 and D2), and a C-terminal extension (CTE) (X. Zhang et al., 2000; DeLaBarre and Brunger, 2003). The NTD consists of a double ψ-barrel followed by a C-terminal four-stranded β-barrel, which is involved in binding VCP cofactors and ubiquitinated substrates (Hänzelmann and Schindelin, 2017). Both D1 and D2 ATPase domains possess a RecA-like subdomain at the N terminus formed by a β sheet of five parallel β strands (β1–5) connected with helices (α1–4) and loops, and an α-helix bundle at the C terminus composed of four or five α helices (α5–8 or α5–9) for D1α and D2α subdomains, respectively. Unlike others in the NSF/Cdc48/Pex AAA+ ATPase family, members of the VCP/Cdc48 subgroup contain a unique CTE that provides the binding interface for a subset of VCP cofactors (Hänzelmann and Schindelin, 2017) but has been found disordered and missing in most resolved structures (X. Zhang et al., 2000; DeLaBarre and Brunger, 2003; Davies et al., 2008; Banerjee et al., 2016; Hänzelmann and Schindelin, 2016). Up till now, multiple structures of VCP in different nucleotide-bound states have been determined; however, our understanding of the VCP apo state remains inconclusive (Xia et al., 2016). Earlier studies revealed a barrel-shaped homohexamer structure of VCP, which can undergo large intrahexameric conformational changes in response to nucleotide binding and hydrolysis (Xia et al., 2016; Schuller et al., 2016). Although the NTDs favor a coplanar “down” position with respect to the D1 domains in the ADP-bound state, they switch to an “up” position above the D1-D2 barrel when D1 is bound with the ATP or its nonhydrolyzable analogs such as ATPγS and AMP-PNP (Banerjee et al., 2016; Schuller et al., 2016; Tang et al., 2010). In addition, the D2 domains undergo a rotational twist with respect to D1 following ATP binding, which is further accompanied by a contraction of the α9 helices and relocation of the CTEs toward the central channel (Banerjee et al., 2016; Hänzelmann and Schindelin, 2016). These conformational changes are functionally important as mutations or ligands perturbing the conformational switches compromise VCP functions (Magnaghi et al., 2013; Niwa et al., 2012; Esaki and Ogura, 2010).
The VCP D1 and D2 AAA domains display marked differences in nucleotide binding and hydrolysis despite their structural similarities. It was found that D1 binds nucleotides much tighter than D2 (Briggs et al., 2008; Chou et al., 2014). Although both D1 and D2 are capable of hydrolyzing ATP, the D2 domain was identified as the major contributor of ATPase activity (Song et al., 2003). On the other hand, ATP hydrolysis in D1 may also be functionally important as mutations in D1 that impair ATP hydrolysis cause cell death (Esaki and Ogura, 2010) and the D1-D2 interdomain communication affects VCP activity (Banerjee et al., 2016; Chou et al., 2014; Ye et al., 2003). Recent studies demonstrated that the nucleotide binding and hydrolysis-mediated conformational changes in D2 provide the main driving force for the “hand-over-hand” substrate threading and unfolding inside the axial channel of the VCP hexamer, which ultimately leads to substrate degradation by the proteasome (N. O. Bodnar and Rapoport, 2017b; N. Bodnar and Rapoport, 2017a; Twomey et al., 2019; Cooney et al., 2019). This structural model largely explains the involvement of VCP in UPS-mediated polyubiquitinated substrate degradation but not in other VCP-dependent processes such as homotypic membrane fusion, endosomal trafficking, and autophagy (van den Boom and Meyer, 2018; Stach and Freemont, 2017). Intriguingly, it was also reported that chemical inhibition of VCP ATPase activity could not impair all of the VCP functions (Anderson et al., 2015). These observations suggest the existence of additional VCP conformational states in order to accommodate its functional diversity. Unraveling these new structural forms is therefore essential for further understanding of VCP functions.
Although VCP has been predominantly characterized as a homohexamer, existence of additional oligomeric states has been implicated for VCP in response to cofactor binding, posttranslational modification and cellular stress (Orme and Bogan, 2012; Cloutier et al., 2013; Wang et al., 2016; Thoms, 2002; Her et al., 2016). To this end, observations of a potential dodecamer state of VCP have been reported (Niwa et al., 2012; Peters et al., 1992) and similar dodecamer particles were identified for VCP's archaeal homolog VAT (Pamnani et al., 1997). Nonetheless, the inherently low abundance and paucity of knowledge on the dodecamer species have hampered further analyses, and the precise mechanism controlling VCP dodecamerization has remained unknown. In this study, we have determined two VCP dodecamer structures using single particle cryo-electron microscopy (cryo-EM) with samples directly purified from mammalian cells or E. coli and elucidated the main structural features of this understudied VCP state, including the apo nucleotide status, the “up”-rotated NTDs, and the α9 helix and CTE-mediated dodecamer assembly. We further uncovered that VCP dodecamer formation is promoted by the apo status of D2. These results bring to light a new regulatory mechanism for VCP oligomerization and expand our understanding on the conformational changes of VCP imparted by its nucleotide status.
Results
Cryo-electron microscopy structural determination of VCP dodecamer
Previous structural and biochemical studies were predominantly carried out with recombinant VCP produced in E. coli. To capture additional states of VCP, we analyzed N-terminal FLAG-tagged VCP expressed in H1299 cells and observed a notable fraction of stable VCP dodecamers in addition to hexamers. As revealed by the blue native PAGE, recombinant VCP purified from H1299 cells yielded two major bands including one corresponding to the hexamer and the other a potentially higher oligomeric state (Figures S1A–S1C). Similar observations were also made for VCP transiently or stably expressed with either FLAG or mCherry tags in the U2OS cells (Figures S1D–S1F). Consistent with results obtained with the N-terminal tagged VCP, endogenous VCP was also found in both hexamer and a higher oligomeric state (Figure S1G), which rules out any potential artifacts due to N-terminal tagging. Further analysis with cryo-EM single particle 2D classification (Figures S1H and S1I) revealed 2D class averages of the VCP dodecamers akin to previous observations (Peters et al., 1992; Pamnani et al., 1997), indicating the presence of two oligomer populations, namely, hexamer and dodecamer, in VCP samples isolated from mammalian cells. To better understand the dodecamer state of VCP, we set out to determine the structure of the dodecamer and to elucidate its structural features and mechanism regulating VCP dodecamerization. A cryo-EM dataset of recombinant VCP purified from H1299 cells was therefore collected and processed (Table 1, Figure S2), from which a dodecamer structure of VCP with an overall resolution of 3.9 Å was resolved. The VCP dodecamer is composed of two hexamers assembled in a tail-to-tail mode, where the D2 domains and CTE tails provide the bulk of the hexamer-hexamer stacking interfaces (Figures 1A and 1B and Video S1).
Table 1.
Cryo-EM data collection, processing, and modeling
| Data collection and processing | |||
| Sample | FLAG-VCP (H1299 cells) | His-VCP (E. coli BL21(DE3)), apyrase treated | |
| Microscope | FEI TITAN KROIS | FEI TITAN KROIS | |
| Imaging mode | Microprobe | Microprobe (EF) | |
| Magnification | 22,500 | 105,000 | |
| Voltage (kV) | 300 | 300 | |
| Total dose (e/Å2) | 46 | 40 | |
| Pixel size (Å/pixel) | 0.65 (super) | 0.69 (super) | |
| Frame rate (frames/second) | 5 | 5 | |
| No. of movies | 618 | 807 | |
| 3D classes | dodecamer | hexamer | dodecamer |
| No. of particles | 27,516 | 22,172 | 38,312 |
| Symmetry imposed | D6 | C6 | D6 |
| Map resolution (Å) | 3.9 | 4.2 | 3.7 |
| Map resolution range (Å) | 3.5–12 | 3.5–11 | 3.5–11 |
| FSC threshold | 0.143 | 0.143 | |
| Refinement | |||
| Initial model used | PDB: 5FTN,5FTK | ||
| Map sharpening B factor (Å2) | −143 | −158 | −139 |
| Model composition | |||
| Protein (residues) | 8,892 | 4,446 | 8,892 |
| Nucleotide | 0 | 0 | 0 |
| RMSD from ideal geometry | |||
| Bonds length (Å) | 0.008 | 0.006 | 0.007 |
| Bonds angle (°) | 1.058 | 1.068 | 1.014 |
| Validation | |||
| Molprobity score | 1.85 | 2.00 | 1.73 |
| Clash score | 7.13 | 6.36 | 6.07 |
| Rotamers outliers (%) | 1.10 | 2.19 | 0.94 |
| Ramachandran plot statistics | |||
| Favored (%) | 93.49 | 94.17 | 94.14 |
| Allowed (%) | 6.51 | 5.83 | 5.86 |
| Disallowed (%) | 0 | 0 | 0 |
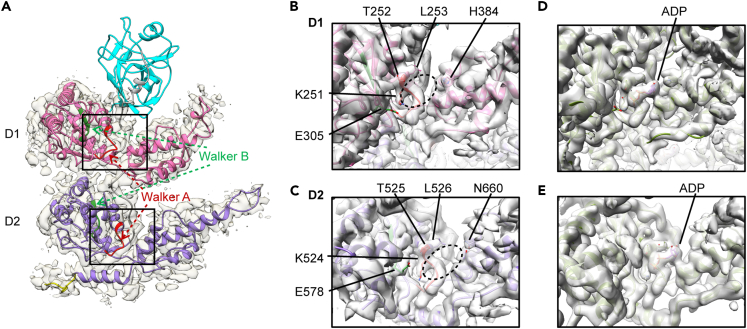
Figure 1.
The 3.9-Å cryo-EM structure of VCP dodecamer purified from mammalian cells
(A) Top view of VCP dodecamer along the central channel.
(B) Side view of VCP dodecamer.
(C) The domain organization and key structural elements of VCP. Each VCP monomer comprises a regulatory N-terminal domain (NTD, residues 1-187), two AAA+ ATPase domains (D1: residues 209–462 and D2: residues 481–762) and a C-terminal extension (CTE, residues 763–806). Both D1 and D2 domains contain Walker A and Walker B motifs. Residues 586–598 in D2 were disordered.
(D) Atomic model of VCP dodecamer of 12 chains (chain A-L) with NTD domains colored in cyan, D1 in pink, D2 in purple, and CTEs in yellow.
(E) CTE in VCP dodecamer.
See also Figures S1 and S2, Videos S1 and S2.
Based on the density map, an atomic model was built for the VCP dodecamer (Figures 1C and 1D). Residues 1–21 and 586–598 were removed from the model due to weak electron densities (Figures S3A and S3B). Comparison of the VCP dodecamer structure with those of the nucleotide-bound hexamers (Banerjee et al., 2016) revealed that the NTDs in the dodecamer are situated in an upward elevated position, similar to those in the hexamer when D1 is occupied by ATPγS. (Figures S3C, S3D, and S4). Consistently, the N-D1 linker in the dodecamer was also found in a similar conformation as that in the D1 ATPγS-bound VCP hexamer (Figure S3E). Structures of D1 and D2 AAA domains in the dodecamer resemble those observed in the hexamers (X. Zhang et al., 2000; DeLaBarre and Brunger, 2003; Davies et al., 2008; Banerjee et al., 2016; Hänzelmann and Schindelin, 2016). Differing from most previously reported VCP structures that usually miss residues after Q763, the CTEs within the dodecamer were largely stabilized and residues were traced up to S775 (Figures 1E and S3A). Binding of ATPγS to the D2 domains of VCP hexamer was also found capable of stabilizing the CTEs (up to F768) but in a conformation distinct from that observed in VCP dodecamer (Figures S3D and S3F). Furthermore, the C-terminal distal α9 helices of D2 and the following CTEs bridge interactions between the two hexamers for dodecamerization (Figures 1D and 1E and Video S2).
The VCP dodecamer is resolved in the apo state
VCP undergoes substantial conformational changes across all domains during nucleotide binding and ATP hydrolysis (Banerjee et al., 2016; Hänzelmann and Schindelin, 2016; Schuller et al., 2016). To better understand the structural features and mechanism of VCP dodecamerization, we therefore examined the potential nucleotide-binding status of the VCP dodecamer. Both D1 and D2 AAA modules contain nucleotide-binding pockets featured with Walker A (Gx(4)GK[TS]) and Walker B (hhhhDE) motifs as well as multiple conserved residues (Figure 2A). Inspection of the nucleotide-binding pockets of the resolved VCP dodecamer density and atomic model revealed no extra densities for potential bound nucleotides in either D1 or D2 AAA modules (Figures 2B and 2C). This is in striking contrast to previous observations with recombinant VCP purified from E. coli, which has been consistently found prebound with ADP (Tang and Xia, 2016; Banerjee et al., 2016; Briggs et al., 2008). To further confirm the nucleotide status of the VCP dodecamer structure, we also lowpass filtered a previously published cryo-EM density map of ADP-bound VCP hexamer to similar resolution for comparison. Corresponding densities arising from the bound ADP in the nucleotide-bound hexameric VCP were missing in the density map of VCP dodecamer, while the rest of the density features matched well between the hexamer and dodecamer structures (Figures 2D and 2E). Collectively, these observations demonstrated that the resolved VCP dodecamer was captured in an apo nucleotide state, a status that is much less understood in comparison with other nucleotide-bound states due to the dominant presence of prebound ADP in most VCP preparations (X. Zhang et al., 2000; DeLaBarre and Brunger, 2003; Banerjee et al., 2016; Hänzelmann and Schindelin, 2016).
Figure 2.
VCP dodecamer was in apo nucleotide status
(A) A single subunit of VCP dodecamer showing nucleotide-binding pockets of D1 and D2. Domains are colored as in Figure 1.
(B) The D1 nucleotide-binding cavity of VCP dodecamer.
(C) The D2 nucleotide-binding cavity of VCP dodecamer. The Walker A and B motifs and key resides in D1 and D2 nucleotide binding cavity are labeled. The dashed ovals indicate potential locations of bound nucleotides.
(D) The D1 nucleotide-binding cavity of ADP-bound VCP hexamer (EMD: 3296, PDB: 5FTK).
(E) The D2 nucleotide-binding cavity of ADP-bound VCP hexamer (EMD: 3296, PDB: 5FTK).
See also Figures S3 and S4.
Comparison of subunits in the apo dodecamer structure with those of different nucleotide-bound hexamers can identify potential conformation changes in VCP following ADP dissociation and ATP binding. Subunits of the apo dodecamer differs significantly from those of the ADP (D1)-bound hexamer in both the NTD and D1α domains (Figure S3C), suggesting that ADP dissociation from D1 can initiate profound conformational changes in D1 and NTD. In contrast, no obvious changes occur following ATPγS binding to the apo D1 (Figure S3D). These observations therefore suggest that dissociation of ADP from D1 and transition to the apo status could prime VCP for ATP binding to D1. On the other hand, only subtle rotations were observed in D2 between the ADP-bound and the apo VCP, suggesting that the observed larger-scale D2 rotations, α9 helices contraction, and relocation of CTEs toward the central channel in the ATPγS (D2)-bound VCP (Banerjee et al., 2016; Hänzelmann and Schindelin, 2016) (Figures S3D and S3F) occur simultaneously upon ATPγS binding. The analysis reveals potentially different prerequisites for ATP binding in D1 and D2 domains, respectively, which may be linked to their distinct properties in nucleotide binding and hydrolysis (Briggs et al., 2008; Chou et al., 2014; Song et al., 2003).
Nucleotide depletion triggers VCP dodecamerization
We next sought to elucidate the mechanism regulating VCP dodecamerization. Given the observed apo nucleotide status and structural changes in VCP dodecamer relative to the nucleotide-bound hexamers (Figures 2 and S3), we hypothesized that loss of nucleotide from the hexamer may trigger VCP dodecamerization. To test this hypothesis, we sought to reconstitute the apo state from recombinant VCP purified from E. coli, which is prebound with ADP and has been used for most previous structural studies (X. Zhang et al., 2000; DeLaBarre and Brunger, 2003; Davies et al., 2008; Banerjee et al., 2016; Hänzelmann and Schindelin, 2016; Schuller et al., 2016). To generate the apo state, the bacterially expressed recombinant VCP was treated with apyrase (Smith et al., 2002), which converts ADP to AMP and has been used previously to produce apo VCP (Davies et al., 2005; Tang and Xia, 2013). In support of our hypothesis, treatment of recombinant VCP by apyrase generated the dodecamer band in blue native PAGE (Figure 3A), similar to that observed for the mammalian samples (Figures S1C–S1G). Negative staining TEM imaging analysis also confirmed the appearance of dodecamer particles from the recombinant VCP sample after apyrase treatment (Figures 3B and 3C). Time course analysis of apyrase treatment revealed an inverse correlation between the level of VCP-bound ADP and the dodecamer/hexamer ratio (Figures S5A and S5B). These results indicate that ADP depletion promotes VCP dodecamer formation. As expected, re-introducing ADP or ATPγS to the apyrase-treated VCP sample converted the dodecamers back to hexamers (Figures S5C–S5F). To ascertain whether the same dodecamer structure resolved for VCP samples purified from mammalian cells is also formed by apyrase-treated recombinant VCP isolated from E. coli, a single particle cryo-EM study was performed (Table 1 and Figure S6). A 3.7-Å structure of bacterial recombinant VCP dodecamer was determined, which was resolved in the apo state as expected (Figure 3D) and found essentially identical to the VCP structure solved from mammalian samples (Figure 3E). Akin to the observations with dodecamers purified from the mammalian cells, this resolved dodecamer structure of apyrase-treated recombinant VCP isolated from E. coli also contained flexible, up-elevated NTDs, as demonstrated by both the D6 and C1 reconstructions (Figures 3D, 3E and S7). In addition, a 4.2-Å apo hexamer structure with similar subunit structural features as the dodecamer was also obtained from the recombinant VCP sample isolated from E. coli (Figure S6A). Taken together, these results demonstrated the linkage of apo nucleotide status of VCP and dodecamerization and confirmed that recombinant VCP isolated from E. coli could form the same dodecamer structure once VCP is in the apo status.
Figure 3.
Apyrase treatment of recombinant VCP purified from E. coli triggered VCP dodecamerization
(A) BN-PAGE of bacterial recombinant His-VCP before and after apyrase treatment to remove prebound ADP. Single and double asterisks mark the band positions of hexamer and dodecamer, respectively.
(B) Negative staining TEM of His-VCP.
(C) Negative staining TEM of His-VCP post apyrase treatment. Zoom-in views of multiple regions show particles resembling dodecamers observed in mammalian samples.
(D) The 3.7-Å cryo-EM structure of apo His-VCP dodecamer obtained by apyrase treatment. The blue lines in D1 and D2 nucleotide-binding cavity composed of Walker A and Walker B motifs mark positions for densities of bound nucleotides that are missing in VCP dodecamer. For clarity, only five chains of the atomic model are displayed.
(E) Structural comparison of the apyrase treatment induced His-VCP dodecamer (colored in cyan) and the VCP dodecamer from mammalian cells as shown in Figure 1 (colored in red).
See also Figures S5, S6, and S7.
Loss of nucleotide binding to D2 AAA module promotes VCP dodecamerization
To further define the mechanism regulating VCP dodecamerization, we generated VCP mutants with impaired ADP binding in either D1 or D2, respectively, and accessed changes in their oligomer states. Key residues forming essential contacts with ADP including K251 and T252 in D1 and K524, T525, and L526 in D2 were mutated (Briggs et al., 2008) (Figures 4A and 4B). Mutations of Thr525 to Val and Leu526 to Ala broke a key hydrogen bond and hydrophobic interaction with ADP, respectively, and caused a substantial enrichment of the dodecamer population for these D2 mutants (Figure 4C). In contrast, no significant increase in dodecamers was observed for the D1 mutants K251A and T252A (Figure 4C). Moreover, dodecamers of the VCP ADP binding deficient mutants can be converted back to hexamers after addition of excess amounts of ADP (Figure 4D). These observations are consistent with the importance of VCP nucleotide occupancy in controlling the hexamer-dodecamer equilibrium (Figures S5C–S5F) and indicate that apo D2 likely plays a major role in promoting VCP dodecamerization. In further support of the important role of apo D2 in triggering VCP dodecamerization, we showed that a recently reported A530T mutant with markedly reduced nucleotide binding in D2 (Her et al., 2016) also exhibited an increased propensity of dodecamer formation in comparison with the wild-type VCP (Figures 4E and 4F), which could be further enhanced by ADP depletion with apyrase. In addition, during the preparation of this article, a dodecamer structure was reported for VCP immobilized with an inhibitor-decorated cryo-EM support (Hoq et al., 2021), which as revealed by the comparison in Figure S8 also contains apo D2. This again validates our conclusion.
Figure 4.
Apo D2 promotes VCP dodecamerization
(A) Structure of ADP-bound hexamer (PDB: 5FTK) and contacts of ADP binding in D1.
(B) Contacts of ADP binding in D2 of ADP-bound hexamer (PDB: 5FTK). Dashed lines indicate hydrogen bonds.
(C) BN-PAGE of bacterial recombinant wild-type (WT) and mutant VCPs with mutations in D1 or D2 defective in ADP binding.
(D) BN-PAGE of WT and mutant VCP in the presence of 5 mM ADP.
(E) BN-PAGE of WT and A530T mutant bacterial recombinant VCP with/without apyrase treatment. Single and double asterisks mark the band positions of hexamer and dodecamer, respectively.
(F) Negative staining TEM of bacterial recombinant A530T. Red arrowheads point to dodecamer particles.
(G) A model for nucleotide status governed conformational and oligomeric changes of VCP. The nucleotide status of D1 primarily controls the conformation of NTD, whereas that of D2 regulates the oligomeric states. Dissociation of ADP from D1 releases the “down” conformation of NTD and switches to the more flexible “up” conformation that is stabilized by ATP binding. On the other hand, evacuation of the D2 nucleotide-binding pockets catalyze dimerization of the hexamers to form dodecamers.
See also Figures S8 and S9.
Unlike T525V and L526A, the K524A mutation, which also significantly attenuated ADP binding in D2 (Briggs et al., 2008; Chou et al., 2014), failed to trigger VCP dodecamerization even after the apyrase treatment (Figures 4C and S9D). This suggests that K524 may serve as an essential residue mediating VCP dodecamerization. As revealed by structural comparisons between the apo dodecamer and ADP-bound hexamer, dissociation of ADP from D2 leads to subtle rotations of the D1-D2 linker, the α1-2 helices, D2α and the loop connecting RecA and D2α subdomains, as well as alterations in the conformation and contacts of K524 (Figures S3C and S9A). Shifting from interaction with the β phosphate in the bound ADP, K524 establishes new contacts with G518, P519 in the Walker A loop, and T623 of the loop harboring residues 623–628 in the apo dodecamer structures, which further bridge stronger contacts with the α9 helix via key residues such as Y755 and F758 (Figures S8A and S8B). This interaction network mediated by primary hubs of K524 and Y755 in apo D2 induces slight contraction of the α9 helices toward the central channel and tunes CTEs to favorable conformations for dodecamerization (Figures S3F and S9C). Particularly, a stronger hydrogen bond forms in the apo structure between the hydroxyl group of Y755 in α9 helix and the carbonyl oxygen of N624 in the 623-628aa loop (Figure S9B), which was demonstrated to be essential for VCP dodecamerization (Figure S9D). In addition, manipulation of other interface residues in the α9 helix and the CTE also substantially affected dodecamer formation (Figure S9D), supporting the importance of the α9 helix and CTE in mediating VCP dodecamerization. Collectively, our results suggest that apo D2, with K524 establishing a new network of contacts to modulate conformations of α9 helices and CTEs, promotes VCP hexamer to dodecamer oligomerization.
Discussion
Previous structural studies revealed that VCP undergoes profound conformational changes within the hexamer assembly during nucleotide binding and hydrolysis (Xia et al., 2016; Schuller et al., 2016). In this study, we have elucidated a new VCP oligomeric state regulated by its nucleotide status. Using VCP purified from mammalian cells and the single particle cryo-EM approach, we have structurally characterized an understudied VCP dodecamer state, which was resolved in the apo nucleotide status with both D1 and D2 AAA domains empty. We further determined the contributions of nucleotide-free D1 and D2 to VCP dodecamerization and found that D2 apo status is essential for VCP dodecamer formation. These findings further extend our current understanding on nucleotide-driven conformational changes of VCP (Figure 4G); the nucleotide status of D1 primarily controls the conformation of NTDs, whereas that of D2 affects the conformation of the α9 helices and CTEs and thereby the oligomeric state of VCP. The dodecamer state is likely unique to the VCP/Cdc48 subgroup of the NSF/Cdc48/Pex AAA+ ATPase family given that the CTE, which provides essential dodecamerization interface, is missing for other subgroups.
One prominent feature of existing VCP structures determined from recombinant VCP/Cdc48 produced in E. coli is the presence of prebound ADP. It is generally believed that almost all D1 domains of purified VCP are loaded with ADP even in the absence of ADP during the course of sample purification (DeLaBarre and Brunger, 2003; Briggs et al., 2008; Tang and Xia, 2013). Recent advance in structural characterization of VCP using single particle cryo-EM further suggests that most D2 domains are also bound with ADP post purification from E. coli (Banerjee et al., 2016). Our observation that VCP dodecamer formation requires D2 in the apo state provides an explanation why the dodecamer state has rarely been captured before. Of interest, when expressed and purified from mammalian cells, a notable fraction of VCP dodecamer species was observed (Figure S1), which furnished us the sample for structural determination of a naturally formed VCP dodecamer for the first time. Although the cellular regulatory mechanism(s) of VCP oligomerization remain to be fully established, we speculate that unknown factors such as post-translational modifications or ligands that can reduce or inhibit VCP nucleotide binding may stabilize the dodecamer state of VCP. A recent study reported a similar dodecamer structure of VCP determined using an inhibitor ligand-decorated affinity gird technique of cryo-EM (Hoq et al., 2021). In agreement with our conclusion that apo D2 induces VCP dodecamerization, all of the D2 domains in this dodecamer structure were also found to be free of nucleotide (Figure S8). However, how exactly this inhibitor perturbs VCP conformation, nucleotide binding, and thereby oligomer state was not fully understood as the inhibitor molecule was not resolved in the structure (Hoq et al., 2021).
As mentioned above, VCP has been primarily resolved in various nucleotide-bound states (X. Zhang et al., 2000; DeLaBarre and Brunger, 2003; Davies et al., 2008; Banerjee et al., 2016; Hänzelmann and Schindelin, 2016), whereas the apo states have remained less well defined. An early structural study of murine VCP with X-ray crystallography reported a hexamer structure with apo D2 (Huyton et al., 2003); however, its D2 domains are highly disordered and miss ∼100 residues in the C terminus. Later, Hanzelmann et al. determined the structure of an engineered α9-D4 variant of VCP carrying four mutations in the α9 helix (N750D/R753D/M757D/Q760D), which somehow lost the prebound ADP during crystallization and was resolved as a hexamer with apo D1 and D2 (Hänzelmann and Schindelin, 2016). Contrary to our observation that dissociation of ADP from D1 initiates the up-relocation of NTD domains, the apo α9-D4 VCP variant was determined in the “down” conformation. This discrepancy might be due to artifact of crystal packing since two additional VCP structures with ATPγS bound to D1 from the same study were also resolved with NTDs in the “down” conformation (Hänzelmann and Schindelin, 2016), differing from the “up” conformation of NTDs revealed by a single particle cryo-EM study (Banerjee et al., 2016). Furthermore, structural comparison of the apo α9-D4 and the dodecamer structures reveals a CTE conformation, which is incompatible with the dodecamer assembly, suggesting that these mutations in the α9 helix alter the CTE assembly and inhibit apo VCP dodecamerization. Indeed, we demonstrated that the α9-D4 mutations such as N750D and R753D block VCP dodecamer formation (Figure S8D). This finding explains why hexamer instead of dodecamer assembly was observed for the apo α9-D4 VCP (Hänzelmann and Schindelin, 2016). More recently, Tang and colleagues reported a crystal structure of an NTD-truncated VCP in complex with the CB-5083 inhibitor, which showed the same dodecamer assembly with apo D1 and D2 (Tang et al., 2019). Although no insight was provided for the formation of an apo nucleotide dodecamer state (Tang et al., 2019), it is possible that truncation of the N-terminal domain may reduce ADP binding, leading to dodecamer formation. Another possibility is that binding of CB-5083 may displace ADP in D2 to induce the apo D2 nucleotide status and thereby transition to the dodecamer state. Unlike earlier studies with engineered VCP variants and inconclusive structural characterization of the apo state, here we report apo structures of wild-type VCP and define key structural features for the D1-related relaxation and up-rotation of NTD domains and the D2-controlled oligomer state change. Structural comparison of subunits among the ADP-bound, apo and ATPγS-bound VCP suggests that dissociation of ADP from D1 may prime its ATP binding by initiating conformational changes in D1 and NTD favorable for ATP binding, whereas that in D2 is not required for ATP loading. This, together with the slow off rate for ADP dissociation from VCP (Briggs et al., 2008), may explain the faster ATP turnover and much higher ATPase activity in D2 relative to D1 (Song et al., 2003).
Increasing lines of evidence suggest a sequential ATP hydrolysis model for the D2 domains of VCP hexamer, where ATP hydrolysis in one protomer primes its anticlockwise neighbor for subsequent hydrolysis (Briggs et al., 2008; Twomey et al., 2019; Cooney et al., 2019). However, VCP dodecamer assembly requires most, if not all, D2 domains to be empty in order to avoid steric clashes in α9 helix and C-terminal extension (Figure S8C). Therefore, the dodecamer state is unlikely to be involved in the regular ATP hydrolysis cycle of VCP. Remarkably, ATP hydrolysis-driven protein unfolding and directional threading through the central tunnel have been proposed as the core underlying molecular mechanism responsible for most VCP-related functions (van den Boom and Meyer, 2018; N. Bodnar and Rapoport, 2017a; Barthelme and Sauer, 2016). Given that VCP dodecamer is assembled by two apo VCP hexamers in a tail-to-tail fashion, it is unlikely that the dodecamer state participates in the UPS-mediated polyubiquitinated substrate degradation. What then might be the cellular function for the dodecamer state of VCP, if any? In contrast to the complete substrate threading model proposed for VCP to direct ubiquitinated proteins to the proteasome (van den Boom and Meyer, 2018; N. Bodnar and Rapoport, 2017a; Twomey et al., 2019; Cooney et al., 2019), a partial threading model has also been proposed for some substrates, particularly those destined to be recycled or reused in processes such as membrane fusion (van den Boom and Meyer, 2018; Stach and Freemont, 2017; Huang et al., 2016). The unique structural properties of the dodecamer state suggest that dodecamerization of VCP could fulfill a control mechanism to block complete substrate threading and promote partial segregation of proteins in cellular processes such as membrane fusion. In this context, it is noteworthy that “holding” or partial threading of substrates by VCP seem independent of its ATPase activity (Twomey et al., 2019; Thoms, 2002; Stach and Freemont, 2017). In line with a potential functional role for the dodecamer VCP, cellular expression of the D2 Walker A mutants, which are predicted to compromise both ATPase activity and dodecamer formation, was reported to cause more severe functional defects than the D2 Walker B mutants, which impair only the ATPase activity but not dodecamerization (Figure S9D), even under conditions where more ATPase activity was preserved for the former (Esaki and Ogura, 2010; Hirabayashi et al., 2001; Kobayashi et al., 2002). Alternatively, similar to the proposed formation of dimeric “hibernating” 100S ribosome complexes in suppressing protein synthesis (Trösch and Willmund, 2019), VCP dodecamerization could also serve as a regulatory “hibernation” mechanism to preserve ATP consumption by VCP given its high cellular abundance (Mori-Konya et al., 2009; Stach and Freemont, 2017). Of interest, during the revision of this article, a new study reported that a disease-causing R155H mutation of VCP triggered the formation of a significant dodecamer population (∼40%), although its functional relevance and link to pathogenesis remain unknown (Nandi et al., 2021). Altogether, future studies will be required to further define the potential involvement of VCP dodecamer in mediating VCP's diverse biological functions.
Limitations of the study
VCP has been best known as a homohexamer. However, a dodecamer state has also been observed. In this study, we have elucidated the mechanism controlling the transition of VCP from the hexamer to dodecamer state using single particle cryo-EM and biochemical assays. Since this work has been primarily carried out in vitro with purified VCP protein from mammalian cells or E. coli, further studies will be required to demonstrate the existence of VCP dodecamer inside the cell, identify the cellular regulatory mechanism(s) controlling VCP oligomerization, and establish the biological significance of the VCP dodecamer state. Given that the apo VCP dodecamer is incompatible with the proposed role for VCP in proteasome-mediated polyubiquitinated protein degradation, the work presented here provides a structural framework for studying other VCP-mediated processes.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-Flag | Sigma Aldrich | Cat # F1804; RRID: AB262044 |
| Mouse polyclonal anti-VCP | Thermo Fisher | Cat # PA5-22257; RRID: AB11157831 |
| Chemicals, peptides, and recombinant proteins | ||
| 3X Flag peptides | Sigma Aldrich | F4799 |
| Anti-Flag M2 affinity gel | Sigma Aldrich | Cat # A2220; RRID: AB10063035 |
| cOmplete™ Protease Inhibitor Cocktail | Roche | 05 056 489 001 |
| Ni-NTA agarose | Qiagen | 30230 |
| Adenosine 5′-diphosphate sodium salt | Sigma Aldrich | A2754 |
| Adenosine 5′-[γ-thio] triphosphate tetralithium salt | Sigma Aldrich | A1388 |
| Adenosine 5′-triphosphate (ATP) disodium salt hydrate | Sigma Aldrich | FLAAS-10VL |
| Apyrase | NEB | Cat # M0398S |
| DMEM with glucose | Corning Cellgro | 10-013-CV |
| Penicillin-Stremptomycin Solution (100x) | Corning | MT30002CI |
| Fetal Bovine Serum | Gibco | 26400044 |
| Bovine Calf Serum | Thermo Fisher | 16777-204 |
| NativePAGE™ 3 to 12% | Thermo Fisher | BN1003BOX |
| PVDF membrane | BioRad | E2311 |
| Superose 6 Increase 10/300 GL | GE Healthcare | GE29-0915-96 |
| Critical commercial assays | ||
| ADP/ATP Ratio Assay Kit | Sigma Aldrich | MAK135-1KT |
| QuikChange Site-Directed Mutagenesis Kit | Agilent | 200521 |
| Experimental models: Cell lines | ||
| NCI-H1299 | ATCC | Cat # CRL-5803 |
| U2OS | ATCC | Cat# CRL-HTB-96 |
| Bacterial and virus strains | ||
| Escherichia Coli BL21(DE3) | Thermo Fisher | EC0114 |
| Oligonucleotides | ||
| VCP (K251A): Forward: CCTCCTGGAAC AGGAGCGACCCTGATTGCTCG |
Integrated DNA Technologies | N/A |
| VCP (K251A): Reverse: CGAGCAATCAGGG TCGCTCCTGTTCCAGGAGG |
Integrated DNA Technologies | N/A |
| VCP (T252A): Forward: GAACAGGAAAG GCCCTGATTGCTCG |
Integrated DNA Technologies | N/A |
| VCP (T252A): Reverse: CGAGCAATCAGG GCCTTTCCTGTTC |
Integrated DNA Technologies | N/A |
| VCP (K524A): Forward: CCTGGCTGTGGG GCAACTTTGTTGGCC |
Integrated DNA Technologies | N/A |
| VCP (K524A): Reverse: GGCCAACAA AGTTGCCCCACAGCCAGG |
Integrated DNA Technologies | N/A |
| VCP (T525V): Forward: GGCTGTGGG AAAGTGTTGTTGGCCAAAGCC |
Integrated DNA Technologies | N/A |
| VCP (T525V): Reverse: GGCTTTGG CCAACAACACTTTCCCACAGCC |
Integrated DNA Technologies | N/A |
| VCP (L526A): Forward: GTGGGAAA ACTGCGTTGGCCAAAGCC |
Integrated DNA Technologies | N/A |
| VCP (L526A): Reverse: GGCTTTGG CCAACGCAGTTTTCCCAC |
Integrated DNA Technologies | N/A |
| VCP (A530T): Forward: GTTGGCCA AAACCATTGCTAATGAATGCCAGG |
Integrated DNA Technologies | N/A |
| VCP (A530T): Reverse: CCTGGCATT CATTAGCAATGGTTTTGGCCAAC |
Integrated DNA Technologies | N/A |
| VCP (Y755A): Forward: CAATGACATTC GGAAGGCTGAGATGTTTGCCCAGACC |
Integrated DNA Technologies | N/A |
| VCP (Y755A): Reverse: GGTCTGGGCAA ACATCTCAGCCTTCCGAATGTCATTG |
Integrated DNA Technologies | N/A |
| VCP (Y755F): Forward: CAATGACATTCG GAAGTTTGAGATGTTTGCCCAGACC |
Integrated DNA Technologies | N/A |
| VCP (Y755F): Reverse: GGTCTGGGCA AACATCTCAAACTTCCGAATGTCATTG |
Integrated DNA Technologies | N/A |
| VCP (N750D): Forward: CCGTTCTGTCA GTGACGATGACATTCGGAAG |
Integrated DNA Technologies | N/A |
| VCP (N750D): Reverse: CTTCCGAATGT CATCGTCACTGACAGAACGG |
Integrated DNA Technologies | N/A |
| VCP (R753D): Forward: CTGTCAGTGACAATGA CATTGACAAGTATGAGATGTTTGC |
Integrated DNA Technologies | N/A |
| VCP (R753D): Reverse: GCAAACATCTCATAC TTGTCAATGTCATTGTCACTGACAG |
Integrated DNA Technologies | N/A |
| VCP (1-775): Forward: GATTCCCTTCATAGAACCAGGGTG |
Integrated DNA Technologies | N/A |
| VCP (1-775): Reverse: CACCCTGGTTCTATGAAGGGAATC |
Integrated DNA Technologies | N/A |
| Recombinant DNA | ||
| pCMV-VCP | This paper | N/A |
| pmCherry-C1-VCP | This paper | N/A |
| PET28a-VCP | This paper | N/A |
| PET28a-VCP (1-775) | This paper | N/A |
| PET28a-VCP (1-761) | This paper | N/A |
| PET28a-VCP (K251A) | This paper | N/A |
| PET28a-VCP (T252A) | This paper | N/A |
| PET28a-VCP (K524A) | This paper | N/A |
| PET28a-VCP (T525V) | This paper | N/A |
| PET28a-VCP (L526A) | This paper | N/A |
| PET28a-VCP (A530T) | This paper | N/A |
| PET28a-VCP (E305Q) | This paper | N/A |
| PET28a-VCP (E578Q) | This paper | N/A |
| PET28a-VCP (Y755F) | This paper | N/A |
| PET28a-VCP (Y755A) | This paper | N/A |
| PET28a-VCP (N750D) | This paper | N/A |
| PET28a-VCP (R753D) | This paper | N/A |
| Software and algorithms | ||
| MotionCor2 | Zheng et al. (2017) | https://emcore.ucsf.edu/ucsf-software |
| Gctf | Zhang (2016) | https://www2.mrc-lmb.cam.ac.uk/research/locally-developed-software/zhang-software/#gctf |
| Gautomatch | Zhang lab | http://www.mrc-lmb.cam.ac.uk/kzhang/ |
| cryoSPARC v2 | Punjani et al. (2017) | https://cryosparc.com/ |
| Coot | Emsley et al. (2010) | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| Phenix | Afonine et al. (2018) | https://phenix-online.org/documentation/reference/real_space_refine.html |
| UCSF Chimera | Pettersen et al. (2004) | https://www.cgl.ucsf.edu/chimera/ |
| Deposited data | ||
| Flag-VCP (H1299 cells), dodecamer state, cryo-EM map | This paper | EMD: 22675 |
| Flag-VCP (H1299 cells), dodecamer state, coordinates | This paper | PDB: 7K56 |
| His-VCP (E. coli BL21(DE3)), apyrase treated, dodecamer state, cryo-EM map | This paper | EMD: 22676 |
| His-VCP (E. coli BL21(DE3)), apyrase treated, dodecamer state, coordinates | This paper | PDB: 7K57 |
| His-VCP (E. coli BL21(DE3)), apyrase treated, hexamer state, cryo-EM map | This paper | EMD: 22678 |
| His-VCP (E. coli BL21(DE3)), apyrase treated, hexamer state, coordinates | This paper | PDB: 7K59 |
| Others | ||
| TED PELLA Lacey Carbon, 300 mesh | TED PELLA, INC. | 01895-F |
| Graphene oxide | Sigma Aldrich | 763705-25ML |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Zhong-Yin Zhang (zhang-zy@purdue.edu).
Materials availability
All unique reagents generated in this study will be shared by the lead contact upon request.
Method details
Cloning and mutagenesis
The full-length sequence of human VCP (1-806 aa) was amplified by PCR with gene-specific primers and then cloned into pCMV or pmCherry-C1 vectors (Addgene) to generate constructs with an N-terminal Flag or mCherry tag for mammalian expression or cloned into pET28a (Novagen) for a construct in frame with a N-terminal hexa-his tag to express in E. coli. Mutations or C-terminal truncations were all generated by PCR site-directed QuickChange mutagenesis.
Cell lines and cell culture
H1299 cells and U2OS cells were used in the study. U2OS cell stably expressing Flag-VCP (U2OS/Flag-VCP) was obtained by co-transfection of pCMV-VCP and PIRES Puro3 (Addgene) plasmids, and selected against puromycin. The mCherry-VCP U2OS stable line (U2OS/mCherry-VCP) was generated by selection under neomycin. All the cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum under a humidified atmosphere containing 5% CO2. For transfection, cells were seeded at 70% confluency in antibiotic-free medium and grown overnight, and then transfected with selected plasmids using polyethylenimine (PEI).
Purification of Flag-VCP from mammalian cells
The pCMV-VCP plasmids were transfected to H1299 cells using PEI and cells were collected after 24-48 hours transient overexpression. U2OS cells stably expressing Flag- or mCheery-VCP were directly cultured and harvested for affinity or gel filtration purification. Cells were lysed with buffer A (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.5% Triton X-100, and 10% glycerol), supplemented with Complete protease inhibitor cocktail tablet (Roche Applied Science). Anti-Flag Affinity Gel (Bimake) was used for affinity purification. After being extensively washed, Flag-VCP was eluted with buffer B (50 mM Tris-HCl pH7.4, 150 mM NaCl,100 μg/ml 3x FLAG peptide (Sigma Aldrich)). The dodecamer species were enriched by collecting earlier elution fractions in gel filtration as shown in Figure S1E. Samples were concentrated and buffer-exchanged to buffer C (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM tris (2-carboxyethyl) phosphine (TCEP)) or buffer D (20 mM Hepes pH7.4, 150 mM NaCl and 1 mM TCEP) using centrifuge filters (Millipore) for SDS-PAGE, blue native PAGE (BN-PAGE, Thermo Fisher) and cryo-EM analyses.
Wild type and mutants His-VCP purification with E. coli
PET28a-VCP wild type and mutants (K251A, T252A, E305Q, R359A, K524A, T525V, L526A, A530T, E578Q, N750D, R753D, Y755F, Y755A, 1-775aa (ΔCTER), 1-761aa (ΔCTE)) were introduced to E. coli BL21(DE3) and protein expression was induced by the addition of 0.4 mM isopropyl β-D-thiogalactopyranoside (IPTG) at 20°C. After 18 hours, cells were harvested for affinity purification with the Ni2+-charged NTA resin (QIAGEN). Briefly, cells were lysed by sonication in buffer E (20 mM Tris-HCl pH7.6, 150 mM NaCl, 5 mM imidazole (IMD) and the Complete protease inhibitor cocktail tablet (Roche Applied Science)). The clarified lysate was used for resin binding and bound protein was eluted with buffer F (20 mM Hepes pH7.4, 50 mM NaCl, 300 mM IMD) after multiple rounds of washing with 20 and 40 mM IMD. The eluted protein was concentrated and buffer-exchanged to final buffer C or D for SDS-PAGE, BN-PAGE, negative staining and cryo-EM analyses. Gel bands brightness quantification was performed in ImageJ.
ADP assay and apyrase treatment of bacterial recombinant His-VCP proteins
An ADP assay kit (Sigma Aldrich) was used to determine the amount of ADP in His-VCP samples and thereby monitor the elimination by apyrase treatment. The measurements were carried out according to the manufacturer's manual. Briefly, His-VCP samples at a concentration of about 1 mg/ml were incubated with the ADP enzyme that converts ADP to ATP, which reacted with D-luciferin in the presence of luciferase to produce illuminance. The illuminance was recorded using a Neo2 multi-mode microplate reader. At least two repeats were performed for each measurement. To remove prebound ADP, bacterial recombinant His-VCP of ∼1-2 mg/ml was treated with apyrase (∼ 0.5U/mg, NEB) at room temperature for hours or shorter incubation time could be used by increasing the working apyrase concentration. Elimination of prebound ADP was confirmed by the ADP assay. After the treatment, apyrase could be removed by repeating the affinity purification with the Ni2+-charged NTA resin (QIAGEN).
Cryo-EM sample preparations and data collection
Purified Flag-VCP dodecamers from the H1299 cells at a concentration of about 0.5 mg/ml in buffer C was applied to a 300-mesh lacy TEM grid (Ted Pella) coated with graphene oxide (Sigma Aldrich), and plunge-frozen using a Cp3 cryo plunger (Gatan). A dataset of ∼600 movies was collected using FEI Titan Krios TEM operating at 300 KV and recorded with the Gatan K2 Summit direct electron detector at super-resolution mode with 225,00× nominal magnification (0.65 Å/super-pixel). A frame rate of 5 frames per second was used. A dose rate of 8 eps and a total exposure of 10 seconds resulted in a total dose of 46 e/Å2. To obtain apo bacterial recombinant His-VCP, sample was treated with apyrase till almost complete elimination of prebound ADP as determined by the ADP assay, re-purified with the Ni2+-charged NTA resin to remove residual apyrase and concentrated to ∼0.5 mg/ml in buffer D for cryo- freezing as described for Flag-VCP. A dataset of ∼800 movies was collected using FEI Titan Krios TEM operating at 300 KV equipped with a GIF-Quantum energy filter with a slit width of 20 eV. Movies were recorded with the Gatan K2 Summit direct electron detector at super-resolution mode with 105,000× nominal magnification (0.69 Å/super-pixel). A frame rate of 5 frames per second was used. A dose rate of 7.6 eps and a total exposure of 8 seconds resulted in a total dose of ∼40 e/Å2.
Image processing
Alignment of frames was performed using MotionCor2 (Zheng et al., 2017). The summed micrographs without dose-weighting (binning 2) were used to determine the contrast transfer function (CTF) parameters with Gctf (K.Zhang, 2016), while the dose-weighted averages were used for particle picking and the rest of image processing. Particles were first picked from a fraction of micrographs using Gautomatch (http://www.mrc-lmb.cam.ac.uk/kzhang/) without a template, followed with a second step of template-based particle picking. The finally picked particles were imported to cyroSPARC v2 (Punjani et al., 2017) for all rest steps of image processing. For Flag-VCP, about 40,000 particles remained after cleaning via 2D classification. Initial models were generated using the ab initio 3D classification in cryoSPARC, and further hetero-refinement revealed a best class of 27,500 dodecamer particles and two other classes of dodecamer or hexamer particles with reduced quality (8,800 and 3,800 particles, respectively). Following 3D homo-refinements of the best class determined a dodecamer structure with a resolution of 3.9 Å. For the apo His-VCP, about 71,600 particles remained after 2D classification cleaning. Initially, ab initio 3D classification in cryoSPARC revealed two 3D classes including a hexamer and a dodecamer. Further hetero-refinement grouped about 38, 300 particles to the hexamer class (class 1), and about 33,300 particles to a dodecamer class (class 2). A second step ab initio 3D classification and hetero-refinement of class 1 resulting in a good hexamer class with about 22,100 particles that were refined to about 4.2 Å after 3D homo-refinement, and a bad hexamer class that was poorly resolved possibly due to extensive conformational heterogeneity or damaged particles. 3D homo-refinement of the dodecamer class with 33,300 particles converged at about 3.7 Å.
Modeling and refinement
For Flag-VCP, the 3.9 Å map of VCP dodecamer was sharpened using phenix.autosharpen and used for modeling. The ATPγS-bound VCP structure (PDB 5FTN) was fitted in the density map as a rigid body using the ‘fit in map’ function in Chimera, and density for a single subunit was segmented using the “color zone” routine in Chimera. Manual adjustment and real-space refinement of the atomic model against the subunit density was performed in COOT (Emsley et al., 2010) to build the initial template. Further refinement was performed in PHENIX using phenix.real_space_refine (Afonine et al., 2018). Due to the poor density quality of the N-terminal domain, residues 22 to 206 were refined as a rigid body. The phenix.map_symmetry and phenix.apply_ncs were used for generating the dodecamer model using the refined single subunit model and the whole density map as inputs, followed by further refinement with phenix.real_space_refine. The quality of the final models was analyzed with MolProbity in Phenix and the refinement statistics were given in Table 1. For His-VCP dodecamer, the 3.7 Å map was used for modeling using the same strategy as the Flag-VCP. To build the atomic model of His-VCP hexamer, the refined single subunit model of His-VCP dodecamer map, was fitted into the 4.2 Å hexamer map, refined using phenix.real_space_refine. The hexamer model was generated with phenix.map_symmetry and phenix.apply_ncs. The quality of the final models was analyzed with MolProbity in Phenix and the refinement statistics were given in Table 1. Chimera software (Pettersen et al., 2004) was used for preparing figures of cryo-EM maps and models.
Negative staining electron microscopy
3 μl of purified VCP samples at a concentration of 0.1 mg/ml was applied to the homemade Formvar-carbon TEM grid (glow discharged) and incubated for 20 seconds. 2% (w/v) uranyl acetate was then used to stain samples on grid for a total time of 1 min. Images were collected using a Tecnai G2 20 transmission electron microscope operated at 200 KV with LaB6 filament.
Gel filtration
The superpose 6 10/300 or increase columns (GE Healthcare) were used for analyzing or purifying VCP samples. Gel filtration was performed in buffer D (20 mM Hepes pH7.4, 150 mM NaCl and 1 mM TCEP). Elution was monitor by absorbance at 280 nm for affinity purified VCP samples, and mCherry fluorescence (Neo2 multi-mode microplate reader) was used to track and determine the elution position of mCherry-VCP in the U2OS/mCherry-VCP stable cell lysates after gel filtration.
Immunoblotting of VCP
For Western blot, purified samples or cell lysates separated by BN-PAGE were transferred to a polyvinylidene difluoride membrane (PVDF, Sigma Aldrich). Anti-VCP/p97 antibody (Thermo Fisher Scientific) was used for primary immunoblotting followed by incubation with the horseradish peroxidase-conjugated secondary antibody (Cell Signaling Technology). The blots were developed by the enhanced chemiluminescence technique (ECL kit, GE Healthcare) and images were collected with an Azure C500 (Azure Biosystems, USA).
Quantification and statistical analysis
This manuscript does not include quantification or statistical analysis.
Acknowledgments
The cryo-EM and negative staining TEM images were collected using the Purdue Cryo-EM facility and Purdue Electron microscopy facility, respectively. The study was supported in part by NIH RO1 CA69202 and the Robert C. and Charlotte Anderson Chair Endowment. The authors also acknowledge NIH P30CA023168 for supporting Purdue University Center for Cancer Research shared resources.
Author contributions
G.Y., Y.B., and Z.-Y.Z. designed the study. G.Y., Y.B., K.L., and O.A. performed experiments. G.Y., Y.B., W.J., and Z.-Y.Z. analyzed the data. G.Y., Y.B., and Z.-Y.Z. wrote the manuscript.
Declaration of interests
The authors claim no competing interests.
Published: November 19, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103310.
Supplemental information
Data and code availability
-
•
Data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
All the cryo-EM density maps and corresponding atomic models generated in this work have been deposited to the EMDB database (EMD: 22675, 22676, 22678) and PDB database (PDB: 7K56, 7K57, 7K59).
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Afonine P.V., Poon B.K., Read R.J., Sobolev O.V., Terwilliger T.C., Urzhumtsev A., Adams P.D. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 2018;74:531–544. doi: 10.1107/S2059798318006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D.J., Le Moigne R., Djakovic S., Kumar B., Rice J., Wong S., Wang J., Yao B., Valle E., Kiss von Soly S. Targeting the AAA ATPase p97 as an approach to treat cancer through disruption of protein homeostasis. Cancer Cell. 2015;28:653–665. doi: 10.1016/j.ccell.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S., Bartesaghi A., Merk A., Rao P., Bulfer S.L., Yan Y., Green N., Mroczkowski B., Neitz R.J., Wipf P. 2.3 Å resolution cryo-EM structure of human p97 and mechanism of allosteric inhibition. Science. 2016;351:871–875. doi: 10.1126/science.aad7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthelme D., Sauer R.T. Origin and functional evolution of the Cdc48/p97/VCP AAA+ protein unfolding and remodeling machine. J. Mol. Biol. 2016;428:1861–1869. doi: 10.1016/j.jmb.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodnar N., Rapoport T. Toward an understanding of the Cdc48/p97 ATPase. F1000Res. 2017;6:1318. doi: 10.12688/f1000research.11683.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodnar N.O., Rapoport T.A. Molecular mechanism of substrate processing by the Cdc48 ATPase complex. Cell. 2017;169:722–735.e9. doi: 10.1016/j.cell.2017.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs L.C., Baldwin G.S., Miyata N., Kondo H., Zhang X., Freemont P.S. Analysis of nucleotide binding to P97 reveals the properties of a tandem AAA hexameric ATPase. J. Biol. Chem. 2008;283:13745–13752. doi: 10.1074/jbc.M709632200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T.-F., Bulfer S.L., Weihl C.C., Li K., Lis L.G., Walters M.A., Schoenen F.J., Lin H.J., Deshaies R.J., Arkin M.R. Specific inhibition of p97/VCP ATPase and kinetic analysis demonstrate interaction between D1 and D2 ATPase domains. J. Mol. Biol. 2014;426:2886–2899. doi: 10.1016/j.jmb.2014.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier P., Lavallée-Adam M., Faubert D., Blanchette M., Coulombe B. A newly uncovered group of distantly related lysine methyltransferases preferentially interact with molecular chaperones to regulate their activity. Plos Genet. 2013;9:e1003210. doi: 10.1371/journal.pgen.1003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney I., Han H., Stewart M.G., Carson R.H., Hansen D.T., Iwasa J.H., Price J.C., Hill C.P., Shen P.S. Structure of the Cdc48 segregase in the act of unfolding an authentic substrate. Science. 2019;365:502–505. doi: 10.1126/science.aax0486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J.M., Tsuruta H., May A.P., Weis W.I. Conformational changes of p97 during nucleotide hydrolysis determined by small-angle X-Ray scattering. Structure. 2005;13:183–195. doi: 10.1016/j.str.2004.11.014. [DOI] [PubMed] [Google Scholar]
- Davies J.M., Brunger A.T., Weis W.I. Improved structures of full-length p97, an AAA ATPase: implications for mechanisms of nucleotide-dependent conformational change. Structure. 2008;16:715–726. doi: 10.1016/j.str.2008.02.010. [DOI] [PubMed] [Google Scholar]
- DeLaBarre B., Brunger A.T. Complete structure of p97/valosin-containing protein reveals communication between nucleotide domains. Nat. Struct. Biol. 2003;10:856–863. doi: 10.1038/nsb972. [DOI] [PubMed] [Google Scholar]
- Deshaies R.J. Proteotoxic crisis, the ubiquitin-proteasome system, and cancer therapy. BMC Biol. 2014;12:94. doi: 10.1186/s12915-014-0094-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esaki M., Ogura T. ATP-bound form of the D1 AAA domain inhibits an essential function of Cdc48p/p97. Biochem. Cell Biol. 2010;88:109–117. doi: 10.1139/o09-116. [DOI] [PubMed] [Google Scholar]
- Her N.-G., Toth J.I., Ma C.-T., Wei Y., Motamedchaboki K., Sergienko E., Petroski M.D. p97 composition changes caused by allosteric inhibition are suppressed by an on-target mechanism that increases the enzyme’s ATPase activity. Cell Chem. Biol. 2016;23:517–528. doi: 10.1016/j.chembiol.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi M., Inoue K., Tanaka K. VCP/p97 in abnormal protein aggregates, cytoplasmic vacuoles, and cell death, phenotypes relevant to neurodegeneration. Cell Death Differ. 2001;8:977–984. doi: 10.1038/sj.cdd.4400907. [DOI] [PubMed] [Google Scholar]
- Hoq M.R., Vago F.S., Li K., Kovaliov M., Nicholas R.J., Huryn D.M., Wipf P., Jiang W., Thompson D.H. Affinity capture of p97 with small-molecule ligand bait reveals a 3.6 Å double-hexamer cryoelectron microscopy structure. ACS Nano. 2021 doi: 10.1021/acsnano.0c10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Tang D., Wang Y. Monoubiquitination of syntaxin 5 regulates golgi membrane dynamics during the cell cycle. Dev. Cell. 2016;38:73–85. doi: 10.1016/j.devcel.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyton T., Pye V.E., Briggs L.C., Flynn T.C., Beuron F., Kondo H., Ma J., Zhang X., Freemont P.S. The crystal structure of murine p97/VCP at 3.6A. J. Struct. Biol. 2003;144:337–348. doi: 10.1016/j.jsb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Hänzelmann P., Schindelin H. Structural basis of ATP hydrolysis and intersubunit signaling in the AAA+ ATPase p97. Structure. 2016;24:127–139. doi: 10.1016/j.str.2015.10.026. [DOI] [PubMed] [Google Scholar]
- Hänzelmann P., Schindelin H. The interplay of cofactor interactions and post-translational modifications in the regulation of the AAA+ ATPase p97. Front. Mol. Biosci. 2017;4:21. doi: 10.3389/fmolb.2017.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Tanaka K., Inoue K., Kakizuka A. Functional ATPase activity of p97/VCP is required for the quality control of endoplasmic reticulum in neuronally differentiated mammalian PC12 cells. J. Biol. Chem. 2002 doi: 10.1074/jbc.M207783200. [DOI] [PubMed] [Google Scholar]
- Magnaghi P., D’Alessio R., Valsasina B., Avanzi N., Rizzi S., Asa D., Gasparri F., Cozzi L., Cucchi U., Orrenius C. Covalent and allosteric inhibitors of the ATPase VCP/p97 induce cancer cell death. Nat. Chem. Biol. 2013;9:548–556. doi: 10.1038/nchembio.1313. [DOI] [PubMed] [Google Scholar]
- Meyer H., Weihl C.C. The VCP/p97 system at a glance: connecting cellular function to disease pathogenesis. J. Cell Sci. 2014;127:3877–3883. doi: 10.1242/jcs.093831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer H., Bug M., Bremer S. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat. Cell Biol. 2012;14:117–123. doi: 10.1038/ncb2407. [DOI] [PubMed] [Google Scholar]
- Mori-Konya C., Kato N., Maeda R., Yasuda K. p97/valosin-containing protein (VCP) is highly modulated by phosphorylation and acetylation. Genes Cell. 2009;14:483–497. doi: 10.1111/j.1365-2443.2009.01286.x. [DOI] [PubMed] [Google Scholar]
- Nandi P., Li S., Columbres R.C.A., Wang F., Williams D.R., Poh Y.-P., Chou T.-F., Chiu P.-L. Structural and functional analysis of disease-linked p97 ATPase mutant complexes. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22158079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H., Ewens C.A., Tsang C., Yeung H.O., Zhang X., Freemont P.S. The role of the N-domain in the ATPase activity of the mammalian AAA ATPase p97/VCP. J. Biol. Chem. 2012;287:8561–8570. doi: 10.1074/jbc.M111.302778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orme C.M., Bogan J.S. The ubiquitin regulatory X (UBX) domain-containing protein TUG regulates the p97 ATPase and resides at the endoplasmic reticulum-golgi intermediate compartment∗. J. Biol. Chem. 2012;287:6679–6692. doi: 10.1074/jbc.M111.284232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamnani V., Tamura T., Lupas A., Peters J., Cejka Z., Ashraf W., Baumeister W. Cloning, sequencing and expression of VAT, a CDC48/p97 ATPase homologue from the archaeon Thermoplasma acidophilum. FEBS Lett. 1997;404:263–268. doi: 10.1016/s0014-5793(97)00138-5. [DOI] [PubMed] [Google Scholar]
- Peters J.-M., Harris J.R., Lustig A., Müller S., Engel A., Volker S., Franke W.W. Ubiquitous soluble Mg2+-ATPase complex: a structural study. J. Mol. Biol. 1992;223:557–571. doi: 10.1016/0022-2836(92)90670-f. [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Schuller J.M., Beck F., Lössl P., Heck A.J.R., Förster F. Nucleotide-dependent conformational changes of the AAA+ ATPase p97 revisited. FEBS Lett. 2016;590:595–604. doi: 10.1002/1873-3468.12091. [DOI] [PubMed] [Google Scholar]
- Smith T.M., Hicks-Berger C.A., Kim S., Kirley T.L. Cloning, expression, and characterization of a soluble calcium-activated nucleotidase, a human enzyme belonging to a new family of extracellular nucleotidases. Arch. Biochem. Biophys. 2002;406:105–115. doi: 10.1016/s0003-9861(02)00420-4. [DOI] [PubMed] [Google Scholar]
- Song C., Wang Q., Li C.-C.H. ATPase activity of p97-valosin-containing protein (VCP). D2 mediates the major enzyme activity, and D1 contributes to the heat-induced activity. J. Biol. Chem. 2003;278:3648–3655. doi: 10.1074/jbc.M208422200. [DOI] [PubMed] [Google Scholar]
- Stach L., Freemont P.S. The AAA+ ATPase p97, a cellular multitool. Biochem. J. 2017;474:2953–2976. doi: 10.1042/BCJ20160783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W.K., Xia D. Altered intersubunit communication is the molecular basis for functional defects of pathogenic p97 mutants. J. Biol. Chem. 2013;288:36624–36635. doi: 10.1074/jbc.M113.488924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W.K., Xia D. Mutations in the human AAA+ chaperone p97 and related diseases. Front. Mol. Biosci. 2016;3:79. doi: 10.3389/fmolb.2016.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W.K., Li D., Li C.-C., Esser L., Dai R., Guo L., Xia D. A novel ATP-dependent conformation in p97 N-D1 fragment revealed by crystal structures of disease-related mutants. EMBO J. 2010;29:2217–2229. doi: 10.1038/emboj.2010.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W.K., Odzorig T., Jin W., Xia D. Structural basis of p97 inhibition by the site-selective anticancer compound CB-5083. Mol. Pharmacol. 2019;95:286–293. doi: 10.1124/mol.118.114256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoms S. Cdc48 can distinguish between native and non-native proteins in the absence of cofactors. FEBS Lett. 2002;520:107–110. doi: 10.1016/s0014-5793(02)02777-1. [DOI] [PubMed] [Google Scholar]
- Trösch R., Willmund F. The conserved theme of ribosome hibernation: from bacteria to chloroplasts of plants. Biol. Chem. 2019;400:879–893. doi: 10.1515/hsz-2018-0436. [DOI] [PubMed] [Google Scholar]
- Twomey E.C., Ji Z., Wales T.E., Bodnar N.O., Ficarro S.B., Marto J.A., Engen J.R., Rapoport T.A. Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding. Science. 2019;365 doi: 10.1126/science.aax1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Boom J., Meyer H. VCP/p97-mediated unfolding as a principle in protein homeostasis and signaling. Mol. Cell. 2018;69:182–194. doi: 10.1016/j.molcel.2017.10.028. [DOI] [PubMed] [Google Scholar]
- Wang T., Xu W., Qin M., Yang Y., Bao P., Shen F., Zhang Z., Xu J. Pathogenic mutations in the valosin-containing protein/p97(VCP) N-domain inhibit the SUMOylation of VCP and lead to impaired stress response. J. Biol. Chem. 2016;291:14373–14384. doi: 10.1074/jbc.M116.729343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia D., Tang W.K., Ye Y. Structure and function of the AAA+ ATPase p97/Cdc48p. Gene. 2016;583:64–77. doi: 10.1016/j.gene.2016.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Meyer H.H., Rapoport T.A. Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J. Cell Biol. 2003;162:71–84. doi: 10.1083/jcb.200302169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Shaw A., Bates P.A., Newman R.H., Gowen B., Orlova E., Gorman M.A., Kondo H., Dokurno P., Lally J. Structure of the AAA ATPase p97. Mol. Cell. 2000;6:1473–1484. doi: 10.1016/s1097-2765(00)00143-x. [DOI] [PubMed] [Google Scholar]
- Zheng S.Q., Palovcak E., Armache J.-P., Verba K.A., Cheng Y., Agard D.A. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017 doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
All the cryo-EM density maps and corresponding atomic models generated in this work have been deposited to the EMDB database (EMD: 22675, 22676, 22678) and PDB database (PDB: 7K56, 7K57, 7K59).
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.