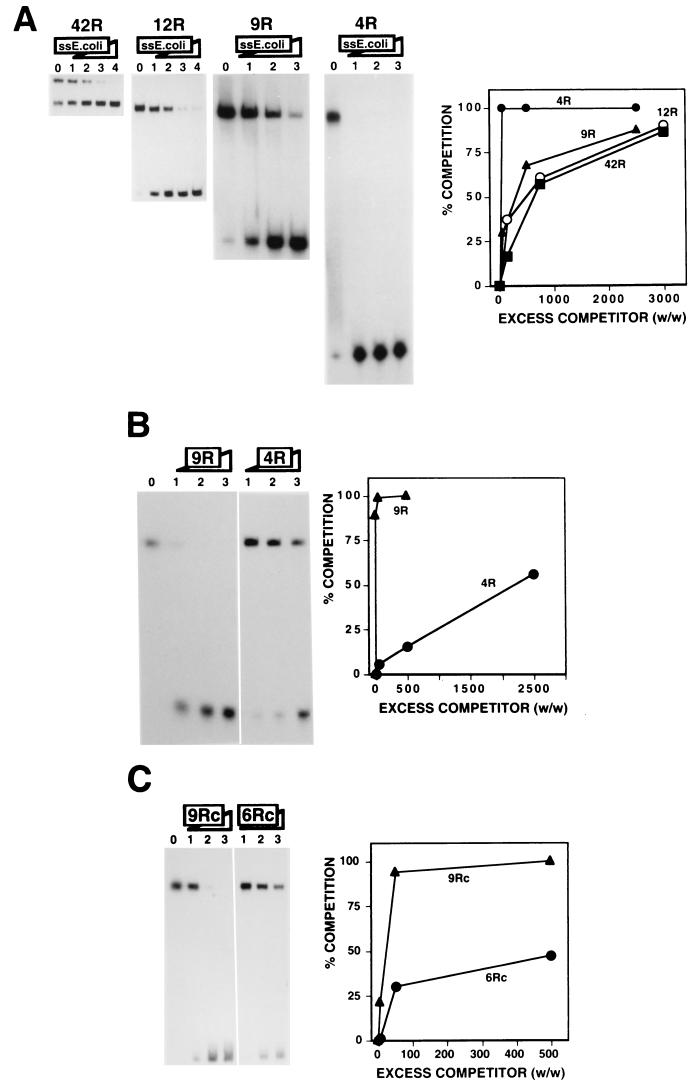
FIG. 1.
Efficient binding of DDP1 to the dodeca-satellite C strand depends on the length of the DNA substrate. (A) The binding of DDP1 to dodeca-satellite C-strand DNA fragments of different lengths is shown as a function of increasing amounts of heat-denatured single-stranded (ss) E. coli DNA. The excess quantities (weight to weight) of competitor used were as follows: panels 42R and 12R, 0 (lanes 0), 150 (lanes 1), 750 (lanes 2), 3,000 (lanes 3), and 9,000 (lanes 4); and panels 9R and 4R, 0 (lanes 0), 50 (lanes 1), 500 (lanes 2), and 2,500 (lanes 3). Quantitative analysis of the results is shown on the right for fragment 42R, fragment 12R, oligo 9R, and oligo 4R. (B) The binding of DDP1 to oligo 9R is shown as a function of increasing amounts of oligo 9R (panel 9R) and oligo 4R (panel 4R). The excess quantities (weight to weight) of competitor used were as follows: panel 9R, 5 (lane 1), 50 (lane 2), and 500 (lane 3); and panel 4R, 50 (lane 1), 500 (lane 2), and 2,500 (lane 3). Lane 0 shows the binding obtained in the absence of any added competitor. Quantitative analysis of the results is shown on the right for oligo 9R and oligo 4R. (C) The binding of DDP1 to oligo 9Rc is shown as a function of increasing excess quantities (weight to weight) of oligo 9Rc (panel 9Rc) and oligo 6Rc (panel 6Rc): 0 (lane 0), 5 (lanes 1), 50 (lanes 2), and 500 (lanes 3). Quantitative analysis of the results is shown on the right for oligo 9Rc and oligo 6Rc. See Table 1 for a description of the DNA fragments used in these experiments.