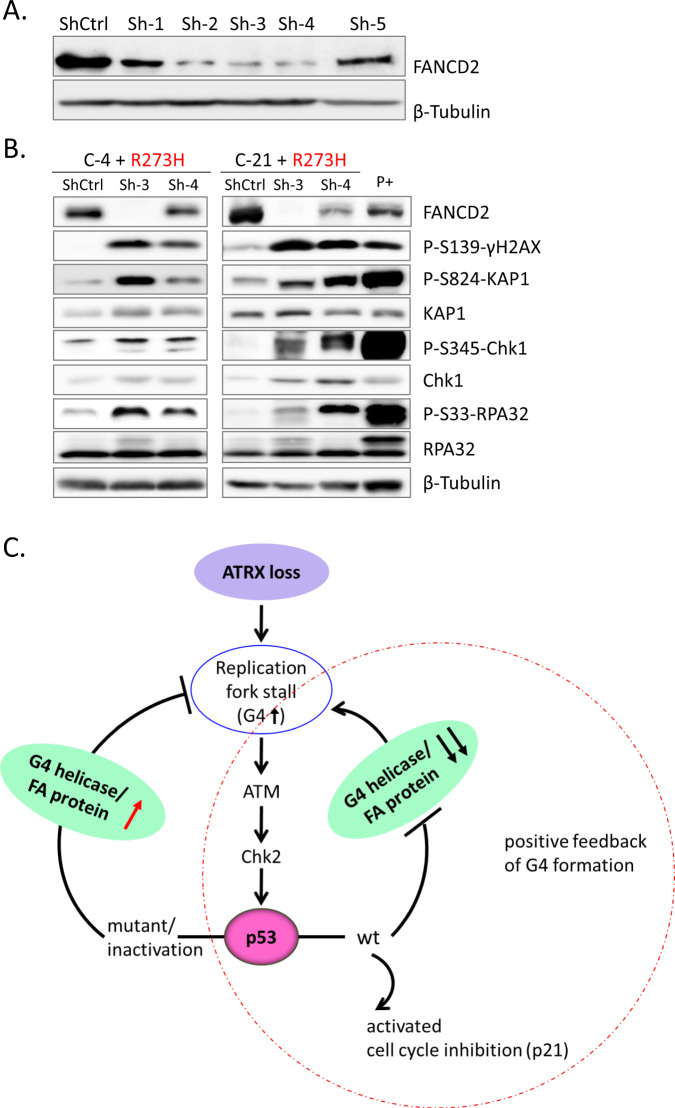
Fig. 6. p53 deficiency limits ATRX loss-induced RS and genome instability through the FA pathway protein, FANCD2.
A A western blot analysis showing the silencing efficiency of shRNAs against FANCD2 in HeLa cells. B FANCD2 protein depletion by shRNA was confirmed by immunoblotting. FANCD2 knockdown in p53-inactivated ATRX KO NGP cells resulted in the further activation of the DDR and RS. NGP cells treated with doxorubicin (0.5 μg/mL, 24 h) were used as a positive control. C Model for linking ATRX, p53, and replication fork-protecting molecules (G4 helicases or the FA pathway protein, FANCD2) to the DDR, RS, and genomic instability. G4 helicases or FANCD2 activation protected cells from RS in p53-inactivated ATRX-deficient cells. In TP53 wt cells, the loss of ATRX resulted in DNA damage and RS in response to replication fork stalling. p53 inactivation inhibited replication fork stalling by triggering G4-resolving helicases and FANCD2 expression, which may promote G4 structure resolution and replication fork protection.